A prototype to process BoxCar data with MSnbase
Source:vignettes/MSnbaseBoxCar.Rmd
MSnbaseBoxCar.RmdAbstract
This package describes a simple prototype to process BoxCar data using the MSnbase package. Is is meant as an illustration of how to useMSnbase to prototype and develop computational mass spectrometry methods and not to replace the reference MaxQuant implementation.
Preparation
Load required packages and functions.
Read a small dataset composed of 16 MS1 spectra as an MSnExp:
f <- dir(system.file("extdata", package = "MSnbaseBoxCar"), pattern = "boxcar.mzML", full.names = TRUE) basename(f)
## [1] "boxcar.mzML"x <- readMSData(f, mode = "onDisk") x
## MSn experiment data ("OnDiskMSnExp")
## Object size in memory: 0.04 Mb
## - - - Spectra data - - -
## MS level(s): 1
## Number of spectra: 16
## MSn retention times: 0:0 - 0:5 minutes
## - - - Processing information - - -
## Data loaded [Sat May 9 18:39:01 2020]
## MSnbase version: 2.15.1
## - - - Meta data - - -
## phenoData
## rowNames: boxcar.mzML
## varLabels: sampleNames
## varMetadata: labelDescription
## Loaded from:
## boxcar.mzML
## protocolData: none
## featureData
## featureNames: F1.S01 F1.S02 ... F1.S16 (16 total)
## fvarLabels: fileIdx spIdx ... spectrum (35 total)
## fvarMetadata: labelDescription
## experimentData: use 'experimentData(object)'Define boxcar groups
Define boxcar groups based on the filterString metadata variable: full scans are encoded as "FTMS + p NSI Full ms [375.0000-1800.0000]" while their respective Boxcar scans reflect the provide adjacent m/z segments "FTMS + p NSI SIM msx ms [299.0000-1701.0000, 299.0000-351.0000, ...]"
fData(x)$filterString[1:4]
## [1] "FTMS + p NSI Full ms [375.0000-1800.0000]"
## [2] "FTMS + p NSI SIM msx ms [299.0000-1701.0000, 299.0000-351.0000, 449.0000-501.0000, 599.0000-651.0000, 749.0000-801.0000, 899.0000-951.0000, 1049.0000-1101.0000, 1199.0000-1251.0000, 1349.0000-1401.0000, 1499.0000-1551.0000, 1649.0000-1701.0000]"
## [3] "FTMS + p NSI SIM msx ms [349.0000-1751.0000, 349.0000-401.0000, 499.0000-551.0000, 649.0000-701.0000, 799.0000-851.0000, 949.0000-1001.0000, 1099.0000-1151.0000, 1249.0000-1301.0000, 1399.0000-1451.0000, 1549.0000-1601.0000, 1699.0000-1751.0000]"
## [4] "FTMS + p NSI SIM msx ms [399.0000-1801.0000, 399.0000-451.0000, 549.0000-601.0000, 699.0000-751.0000, 849.0000-901.0000, 999.0000-1051.0000, 1149.0000-1201.0000, 1299.0000-1351.0000, 1449.0000-1501.0000, 1599.0000-1651.0000, 1749.0000-1801.0000]"The bc_groups function identifies full (noted NA) and BoxCar spectra and groups the latter:
x <- bc_groups(x) fData(x)$bc_groups
## [1] NA 1 1 1 NA 2 2 2 NA 3 3 3 NA 4 4 4Keep only BoxCar spectra
The next filter BoxCar spectra, as defined above.
xbc <- filterBoxCar(x) fData(xbc)$bc_groups
## [1] 1 1 1 2 2 2 3 3 3 4 4 4## Warning: Removed 4448 rows containing missing values (geom_segment).## Warning: Removed 29 rows containing missing values (geom_rect).
Beginning of the first adjacent BoxCar segments.
Combine BoxCar spectra
Remove any peaks outside of the BoxCar segments.
xbc <- bc_zero_out_box(xbc, offset = 0.5) xbc
## MSn experiment data ("MSnExp")
## Object size in memory: 0.37 Mb
## - - - Spectra data - - -
## MS level(s): 1
## Number of spectra: 12
## MSn retention times: 0:0 - 0:5 minutes
## - - - Processing information - - -
## Data converted from Spectra: Sat May 9 18:39:01 2020
## BoxCar processed [Sat May 9 18:39:01 2020]
## MSnbase version: 2.15.1
## - - - Meta data - - -
## phenoData
## rowNames: 1
## varLabels: sampleNames
## varMetadata: labelDescription
## Loaded from:
## 1
## protocolData: none
## featureData
## featureNames: F1.S02 F1.S03 ... F1.S16 (12 total)
## fvarLabels: fileIdx spIdx ... bc_groups (36 total)
## fvarMetadata: labelDescription
## experimentData: use 'experimentData(object)'## Warning: Removed 4448 rows containing missing values (geom_segment).## Warning: Removed 29 rows containing missing values (geom_rect).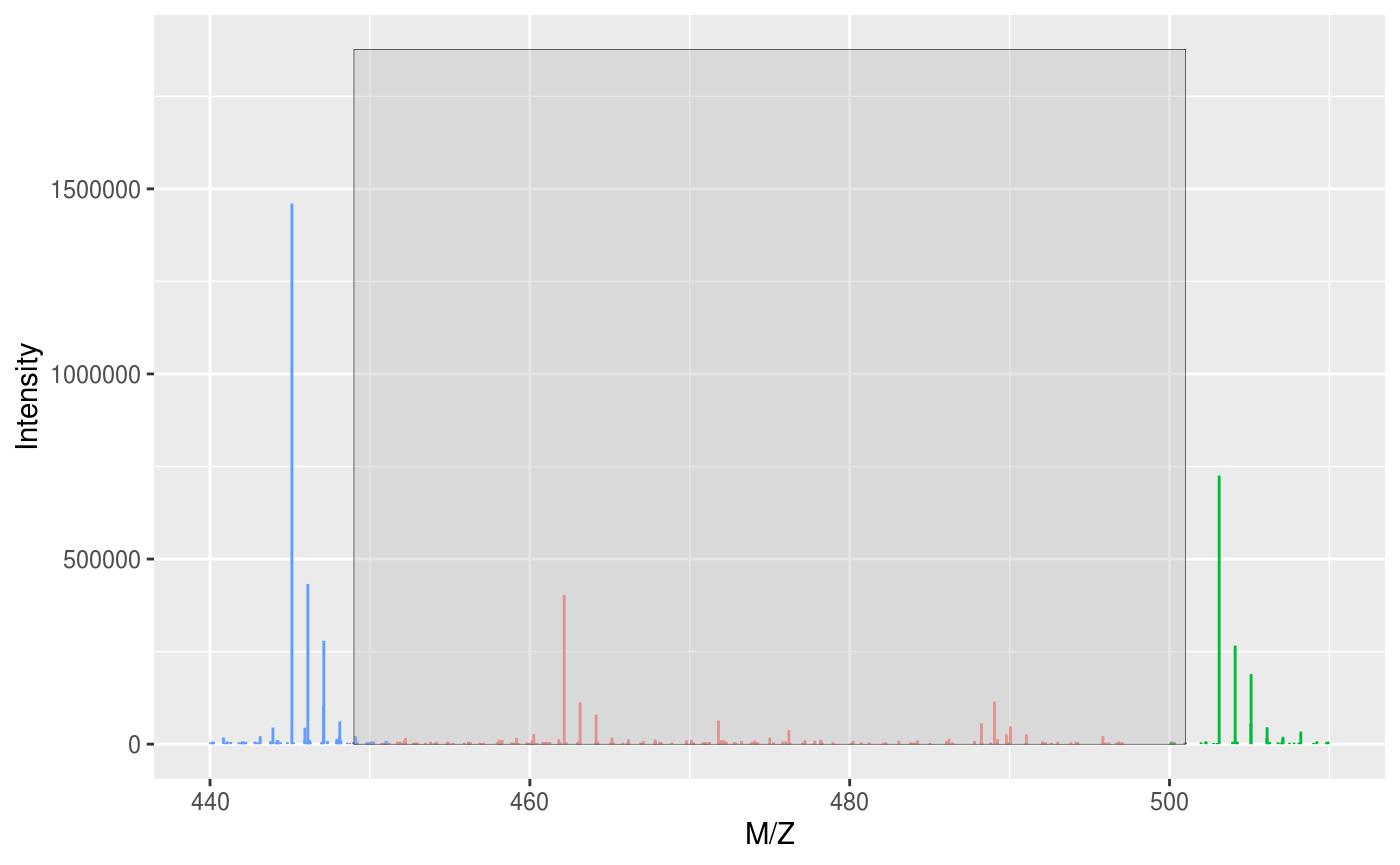
Peaks outside of the BoxCar segments have been removed.
Combine BoxCar spectra to reconstitute the full scan and coerce result back to an MSnExp object containing 4 spectra.
res <- combineSpectra(xbc, fcol = "bc_groups", method = boxcarCombine) res
## MSn experiment data ("MSnExp")
## Object size in memory: 0.34 Mb
## - - - Spectra data - - -
## MS level(s): 1
## Number of spectra: 4
## MSn retention times: 0:0 - 0:4 minutes
## - - - Processing information - - -
## Data converted from Spectra: Sat May 9 18:39:01 2020
## BoxCar processed [Sat May 9 18:39:01 2020]
## Spectra combined based on feature variable 'bc_groups' [Sat May 9 18:39:02 2020]
## MSnbase version: 2.15.1
## - - - Meta data - - -
## phenoData
## rowNames: 1
## varLabels: sampleNames
## varMetadata: labelDescription
## Loaded from:
## 1
## protocolData: none
## featureData
## featureNames: F1.S02 F1.S06 F1.S10 F1.S14
## fvarLabels: fileIdx spIdx ... bc_groups (36 total)
## fvarMetadata: labelDescription
## experimentData: use 'experimentData(object)'plot(res[[1]])

Reconstructed full spectrum.
The above steps can also be piped into a single command.
library("magrittr") res <- x %>% bc_groups() %>% filterBoxCar() %>% bc_zero_out_box(offset = 0.5) %>% combineSpectra(fcol = "groups", method = boxcarCombine)
The processed data can also be written to a new mzML file.
writeMSData(res, "boxcar_processed.mzML")
Session information
## R Under development (unstable) (2020-05-09 r78394)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 14.04.5 LTS
##
## Matrix products: default
## BLAS: /home/travis/R-bin/lib/R/lib/libRblas.so
## LAPACK: /home/travis/R-bin/lib/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 parallel stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] ggplot2_3.3.0 MSnbaseBoxCar_0.2.0 MSnbase_2.15.1
## [4] ProtGenerics_1.21.0 S4Vectors_0.27.5 mzR_2.23.0
## [7] Rcpp_1.0.4.6 Biobase_2.49.0 BiocGenerics_0.35.2
## [10] BiocStyle_2.17.0
##
## loaded via a namespace (and not attached):
## [1] lattice_0.20-41 assertthat_0.2.1 rprojroot_1.3-2
## [4] digest_0.6.25 foreach_1.5.0 R6_2.4.1
## [7] plyr_1.8.6 backports_1.1.6 mzID_1.27.0
## [10] evaluate_0.14 highr_0.8 pillar_1.4.4
## [13] zlibbioc_1.35.0 rlang_0.4.6 preprocessCore_1.51.0
## [16] rmarkdown_2.1 pkgdown_1.5.1 desc_1.2.0
## [19] labeling_0.3 BiocParallel_1.23.0 stringr_1.4.0
## [22] munsell_0.5.0 compiler_4.1.0 xfun_0.13
## [25] pkgconfig_2.0.3 pcaMethods_1.81.0 htmltools_0.4.0
## [28] tidyselect_1.0.0 tibble_3.0.1 bookdown_0.18
## [31] IRanges_2.23.4 codetools_0.2-16 XML_3.99-0.3
## [34] crayon_1.3.4 dplyr_0.8.5 withr_2.2.0
## [37] MASS_7.3-51.6 grid_4.1.0 gtable_0.3.0
## [40] lifecycle_0.2.0 affy_1.67.0 magrittr_1.5
## [43] scales_1.1.0 ncdf4_1.17 stringi_1.4.6
## [46] impute_1.63.0 farver_2.0.3 fs_1.4.1
## [49] affyio_1.59.0 doParallel_1.0.15 limma_3.45.0
## [52] ellipsis_0.3.0 vctrs_0.2.4 iterators_1.0.12
## [55] tools_4.1.0 glue_1.4.0 purrr_0.3.4
## [58] yaml_2.2.1 colorspace_1.4-1 BiocManager_1.30.10
## [61] vsn_3.57.0 MALDIquant_1.19.3 memoise_1.1.0
## [64] knitr_1.28