Chapter 1 Getting started with R and RStudio
Objectives
- Familiarise yourselves with R and RStudio, and how to work with them
- Find help about R
- Install new R packages
- R variables, functions, vectors
- Simple arithmetics and data manipulation
- Learn about R markdown
The first two sections are adapted from the Data Carpentry R for data analysis and visualization material.
1.1 R and RStudio
1.1.1 What is R? What is RStudio?
The term “R” is used to refer to both the programming language and the software that interprets the scripts written using it.
RStudio is currently a very popular way to not only write your R scripts but also to interact with the R software. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
1.1.2 Why learn R?
R does not involve lots of pointing and clicking, and that’s a good thing
The learning curve might be steeper than with other software, but with R, the results of your analysis does not rely on remembering a succession of pointing and clicking, but instead on a series of written commands, and that’s a good thing! So, if you want to redo your analysis because you collected more data, you don’t have to remember which button you clicked in which order to obtain your results, you just have to run your script again.
Working with scripts makes the steps you used in your analysis clear, and the code you write can be inspected by someone else who can give you feedback and spot mistakes.
Working with scripts forces you to have a deeper understanding of what you are doing, and facilitates your learning and comprehension of the methods you use.
R code is great for reproducibility
Reproducibility is when someone else (including your future self) can obtain the same results from the same dataset when using the same analysis.
R integrates with other tools to generate manuscripts from your code. If you collect more data, or fix a mistake in your dataset, the figures and the statistical tests in your manuscript are updated automatically.
An increasing number of journals and funding agencies expect analyses to be reproducible, so knowing R will give you an edge with these requirements.
R is interdisciplinary and extensible
With 10,000+ packages that can be installed to extend its capabilities, R provides a framework that allows you to combine statistical approaches from many scientific disciplines to best suit the analytical framework you need to analyze your data. For instance, R has packages for image analysis, GIS, time series, population genetics, and a lot more.
R works on data of all shapes and sizes
The skills you learn with R scale easily with the size of your dataset. Whether your dataset has hundreds or millions of lines, it won’t make much difference to you.
R is designed for data analysis. It comes with special data structures and data types that make handling of missing data and statistical factors convenient.
R can connect to spreadsheets, databases, and many other data formats, on your computer or on the web.
R produces high-quality graphics
The plotting functionalities in R are endless, and allow you to adjust any aspect of your graph to convey most effectively the message from your data.
R has a large community
Thousands of people use R daily. Many of them are willing to help you through mailing lists and websites such as Stack Overflow.
Not only is R free, but it is also open-source and cross-platform
Anyone can inspect the source code to see how R works. Because of this transparency, there is less chance for mistakes, and if you (or someone else) find some, you can report and fix bugs.
1.1.3 Knowing your way around RStudio
Let’s start by learning about RStudio, which is an Integrated Development Environment (IDE) for working with R.
The RStudio IDE open-source product is free under the Affero General Public License (AGPL) v3. The RStudio IDE is also available with a commercial license and priority email support from RStudio, Inc.
We will use RStudio IDE to write code, navigate the files on our computer, inspect the variables we are going to create, and visualize the plots we will generate. RStudio can also be used for other things (e.g., version control, developing packages, writing Shiny apps) that we will not cover during the workshop.
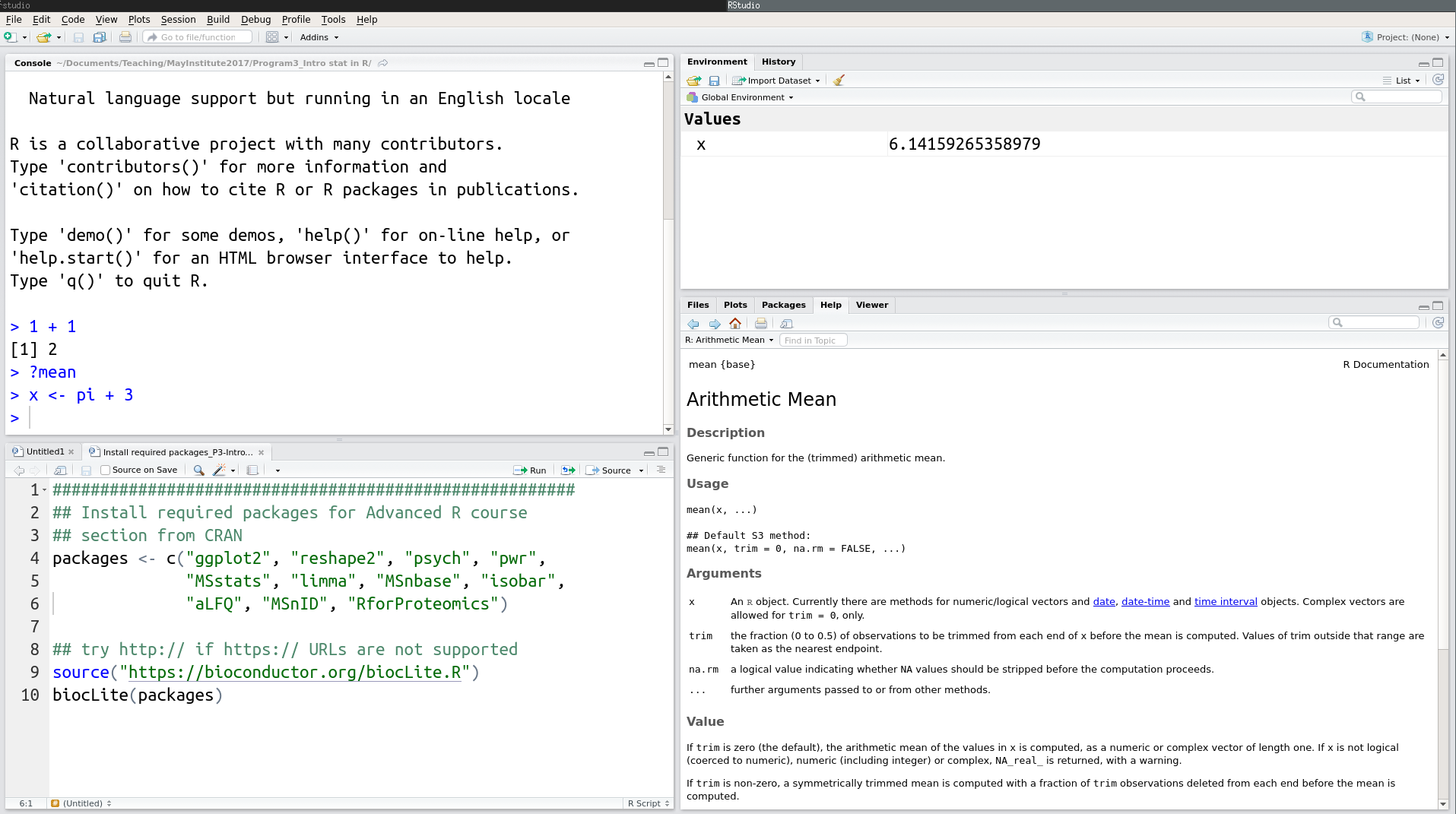
RStudio interface screenshot
RStudio is divided into 4 “Panes”: the Source for your scripts and documents (top-left, in the default layout), the R Console (bottom-left), your Environment/History (top-right), and your Files/Plots/Packages/Help/Viewer (bottom-right). The placement of these panes and their content can be customized (see menu, Tools -> Global Options -> Pane Layout). One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, with many shortcuts, autocompletion, and highlighting for the major file types you use while developing in R, RStudio will make typing easier and less error-prone.
1.1.4 Getting set up
It is good practice to keep a set of related data, analyses, and text self-contained in a single folder, called the working directory. All of the scripts within this folder can then use relative paths to files that indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer). Working this way makes it a lot easier to move your project around on your computer and share it with others without worrying about whether or not the underlying scripts will still work.
RStudio provides a helpful set of tools to do this through its “Projects” interface, which not only creates a working directory for you but also remembers its location (allowing you to quickly navigate to it) and optionally preserves custom settings and open files to make it easier to resume work after a break. Below, we will go through the steps for creating an “R Project” for this tutorial.
- Under the
Filemenu, click onNew project, chooseNew directory, thenEmpty project - Enter a name for this new folder (or “directory”), and choose a convenient location for it. This will be your working directory for the rest of the day (e.g.,
rstats) - Click on
Create project - Under the
Filestab on the right of the screen, click onNew Folderand create a folder nameddatawithin your newly created working directory (e.g.,rstats/data) - Create a new R script in your working directory, calling it e.g.,
rstats-script.R.
Your working directory should now look like this:
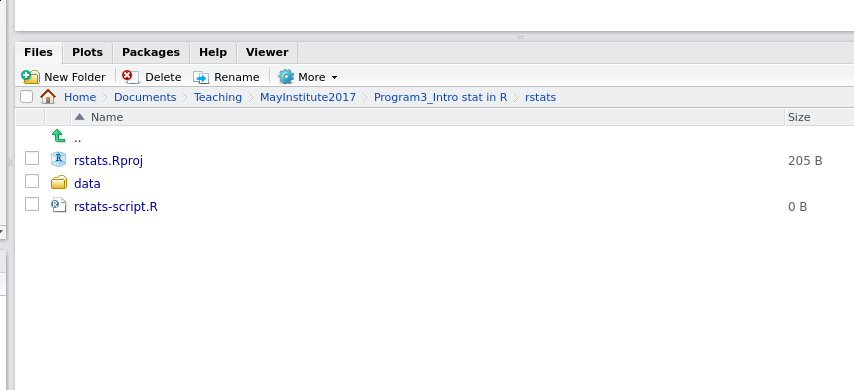
How it should look like at the beginning of this lesson
Challenge
Add the provided data files (
iPRG_example_runsummary.csv,TCGA_sample_information.csv,iprg.rdaandiprg2.rda) to yourdatadirectory.
1.1.5 Organizing your working directory
Using a consistent folder structure across your projects will help keep things organized, and will also make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you may create directories (folders) for scripts, data, and documents.
data/Use this folder to store your raw data and intermediate datasets you may create for the need of a particular analysis. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually) as possible. Separating raw data from processed data is also a good idea. For example, you could have filesdata/raw/prots_plot1.txtand...plot2.txtkept separate from adata/processed/prots.csvfile generated by thescripts/01_preprocess_prots.Rscript.documents/This would be a place to keep outlines, drafts, and other text.scripts/This would be the location to keep your R scripts for different analyses or plotting, and potentially a separate folder for your functions (more on that later).
You may want additional directories or subdirectories depending on your project needs, but these should form the backbone of your working directory. For this workshop, we will need a data/ folder to store our raw data, and we will create later a data_output/ folder when we learn how to export data as CSV files.
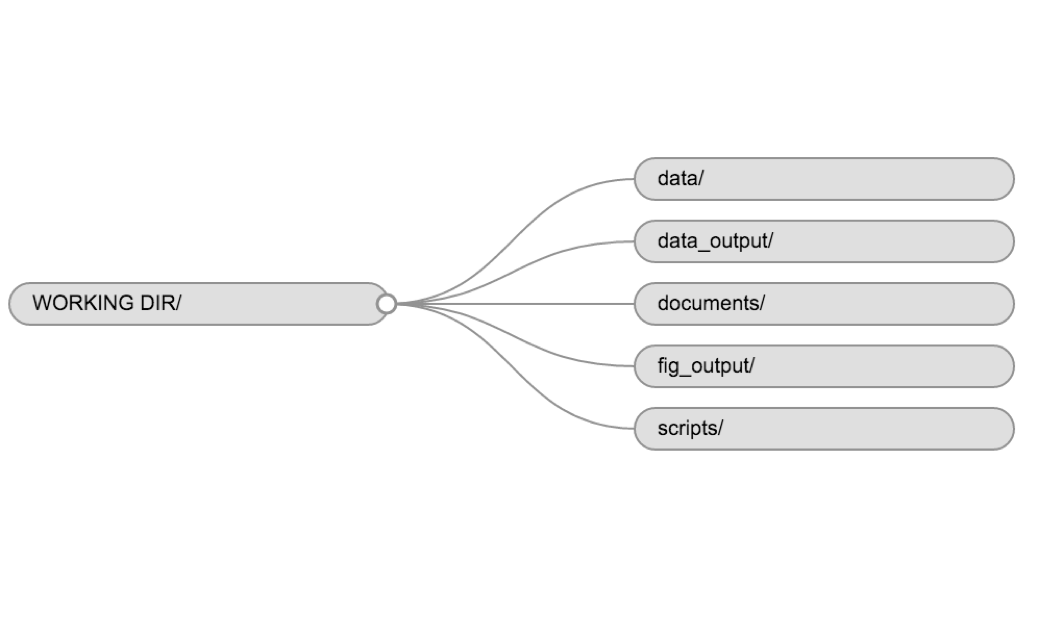
Example of a working directory structure
1.2 Interacting with R
The basis of programming is that we write down instructions for the computer to follow, and then we tell the computer to follow those instructions. We write, or code, instructions in R because it is a common language that both the computer and we can understand. We call the instructions commands and we tell the computer to follow the instructions by executing (also called running) those commands.
There are two main ways of interacting with R: by using the console or by using script files (plain text files that contain your code). The console pane (in RStudio, the bottom left panel) is the place where commands written in the R language can be typed and executed immediately by the computer. It is also where the results will be shown for commands that have been executed. You can type commands directly into the console and press Enter to execute those commands, but they will be forgotten when you close the session.
Because we want our code and workflow to be reproducible, it is better to type the commands we want in the script editor, and save the script. This way, there is a complete record of what we did, and anyone (including our future selves!) can easily replicate the results on their computer.
RStudio allows you to execute commands directly from the script editor by using the Ctrl + Enter shortcut (on Macs, Cmd + Return will work, too). The command on the current line in the script (indicated by the cursor) or all of the commands in the currently selected text will be sent to the console and executed when you press Ctrl + Enter.
At some point in your analysis you may want to check the content of a variable or the structure of an object, without necessarily keeping a record of it in your script. You can type these commands and execute them directly in the console. RStudio provides the Ctrl + 1 and Ctrl + 2 shortcuts allow you to jump between the script and the console panes.
If R is ready to accept commands, the R console shows a > prompt. If it receives a command (by typing, copy-pasting or sent from the script editor using Ctrl + Enter), R will try to execute it, and when ready, will show the results and come back with a new > prompt to wait for new commands.
If R is still waiting for you to enter more data because it isn’t complete yet, the console will show a + prompt. It means that you haven’t finished entering a complete command. This is because you have not ‘closed’ a parenthesis or quotation, i.e. you don’t have the same number of left-parentheses as right-parentheses, or the same number of opening and closing quotation marks. When this happens, and you thought you finished typing your command, click inside the console window and press Esc; this will cancel the incomplete command and return you to the > prompt.
1.3 Seeking help
Use the built-in RStudio help interface to search for more information on R functions
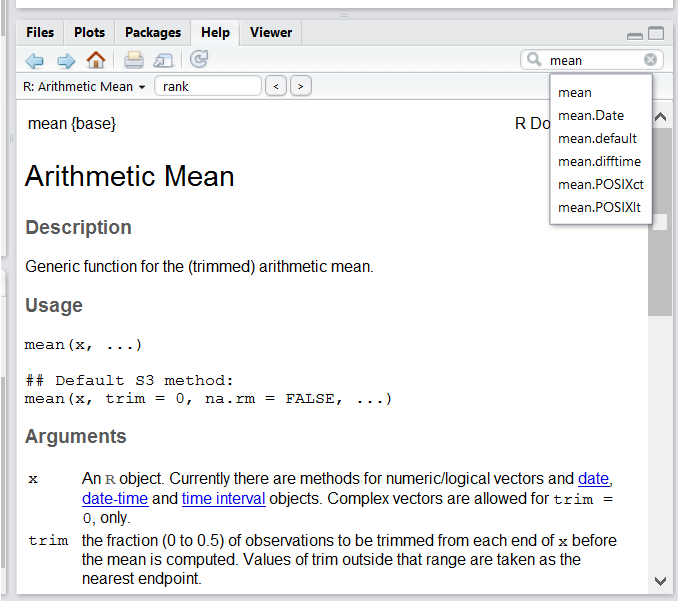
RStudio help interface
One of the most fastest ways to get help, is to use the RStudio help interface. This panel by default can be found at the lower right hand panel of RStudio. As seen in the screenshot, by typing the word “Mean”, RStudio tries to also give a number of suggestions that you might be interested in. The description is then shown in the display window.
I know the name of the function I want to use, but I’m not sure how to use it
If you need help with a specific function, let’s say barplot(), you can type:
?barplotIf you just need to remind yourself of the names of the arguments, you can use:
args(lm)I want to use a function that does X, there must be a function for it but I don’t know which one…
If you are looking for a function to do a particular task, you can use the help.search() function, which is called by the double question mark ??. However, this only looks through the installed packages for help pages with a match to your search request
??kruskalIf you can’t find what you are looking for, you can use the rdocumentation.org website that searches through the help files across all packages available.
Finally, a generic Google or internet search “R <task>” will often either send you to the appropriate package documentation or a helpful forum where someone else has already asked your question.
I am stuck… I get an error message that I don’t understand
Start by googling the error message. However, this doesn’t always work very well because often, package developers rely on the error catching provided by R. You end up with general error messages that might not be very helpful to diagnose a problem (e.g. “subscript out of bounds”). If the message is very generic, you might also include the name of the function or package you’re using in your query.
However, you should check Stack Overflow. Search using the [r] tag. Most questions have already been answered, but the challenge is to use the right words in the search to find the answers: http://stackoverflow.com/questions/tagged/r
The Introduction to R can also be dense for people with little programming experience but it is a good place to understand the underpinnings of the R language.
The R FAQ is dense and technical but it is full of useful information.
Asking for help
The key to receiving help from someone is for them to rapidly grasp your problem. You should make it as easy as possible to pinpoint where the issue might be.
Try to use the correct words to describe your problem. For instance, a package is not the same thing as a library. Most people will understand what you meant, but others have really strong feelings about the difference in meaning. The key point is that it can make things confusing for people trying to help you. Be as precise as possible when describing your problem.
If possible, try to reduce what doesn’t work to a simple reproducible example. If you can reproduce the problem using a very small data frame instead of your 50,000 rows and 10,000 columns one, provide the small one with the description of your problem. When appropriate, try to generalize what you are doing so even people who are not in your field can understand the question. For instance instead of using a subset of your real dataset, create a small (3 columns, 5 rows) generic one. For more information on how to write a reproducible example see this article by Hadley Wickham.
To share an object with someone else, if it’s relatively small, you can use the function dput(). It will output R code that can be used to recreate the exact same object as the one in memory:
## iris is an example data frame that comes with R and head() is a
## function that returns the first part of the data frame
dput(head(iris))## structure(list(Sepal.Length = c(5.1, 4.9, 4.7, 4.6, 5, 5.4),
## Sepal.Width = c(3.5, 3, 3.2, 3.1, 3.6, 3.9), Petal.Length = c(1.4,
## 1.4, 1.3, 1.5, 1.4, 1.7), Petal.Width = c(0.2, 0.2, 0.2,
## 0.2, 0.2, 0.4), Species = structure(c(1L, 1L, 1L, 1L, 1L,
## 1L), .Label = c("setosa", "versicolor", "virginica"), class = "factor")), row.names = c(NA,
## 6L), class = "data.frame")If the object is larger, provide either the raw file (i.e., your CSV file) with your script up to the point of the error (and after removing everything that is not relevant to your issue). Alternatively, in particular if your question is not related to a data frame, you can save any R object to a file using an R-specific binary format:
save(iris, file="iris.rda")The content of this file is however not human readable, but can easily and efficiently be loaded back into R on any other platform with:
load(file="iris.rds")Challenge
Load the
iprg.rdadata into your R session.
Last, but certainly not least, always include the output of sessionInfo() as it provides critical information about your platform, the versions of R and the packages that you are using, and other information that can be very helpful to understand your problem.
sessionInfo()## R version 3.6.0 RC (2019-04-21 r76417)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 18.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/libf77blas.so.3.10.3
## LAPACK: /usr/lib/x86_64-linux-gnu/atlas/liblapack.so.3.10.3
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=fr_FR.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=fr_FR.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## loaded via a namespace (and not attached):
## [1] compiler_3.6.0 magrittr_1.5 bookdown_0.9 htmltools_0.3.6
## [5] tools_3.6.0 rstudioapi_0.10 yaml_2.2.0 Rcpp_1.0.1
## [9] stringi_1.4.3 rmarkdown_1.12 knitr_1.22 stringr_1.4.0
## [13] digest_0.6.18 xfun_0.6 evaluate_0.13Where to ask for help?
- The person sitting next to you during the workshop. Don’t hesitate to talk to your neighbor during the workshop, compare your answers, and ask for help. You might also be interested in organizing regular meetings following the workshop to keep learning from each other.
- Your friendly colleagues: if you know someone with more experience than you, they might be able and willing to help you.
- Stack Overflow: if your question hasn’t been answered before and is well crafted, chances are you will get an answer in less than 5 min. Remember to follow their guidelines on how to ask a good question.
- The R-help mailing list: it is read by a lot of people (including most of the R core team), a lot of people post to it, but the tone can be pretty dry, and it is not always very welcoming to new users. If your question is valid, you are likely to get an answer very fast but don’t expect that it will come with smiley faces. Also, here more than anywhere else, be sure to use correct vocabulary (otherwise you might get an answer pointing to the misuse of your words rather than answering your question). You will also have more success if your question is about a base function rather than a specific package.
- If your question is about a specific package, see if there is a mailing list for it. Usually it’s included in the DESCRIPTION file of the package that can be accessed using
packageDescription("name-of-package"). You may also want to try to email the author of the package directly, or open an issue on the code repository (e.g., GitHub). - There are also some topic-specific mailing lists (GIS, phylogenetics, etc…), the complete list is here.
- If you use any Bioconductor, for example any of the proteomics, metabolomics or mass-spectrometry packages, the support forum is the best place to contact the package author and the wide community.
1.3.1 More resources
- The Posting Guide for the R mailing lists.
- How to ask for R help useful guidelines
- This blog post by Jon Skeet has quite comprehensive advice on how to ask programming questions.
- The reprex package is very helpful to create reproducible examples when asking for help.
1.4 Installing packages
To install a new package, using the install.packages function:
install.package("ggplot2")Bioconductor (and CRAN) packages can be installed with:
install.packages("BiocManager") ## only once
BiocManager::install("MSnbase")1.5 Introduction to R
1.5.1 Creating objects in R
You can get output from R simply by typing math in the console:
3 + 5## [1] 812 / 7## [1] 1.714286However, to do useful and interesting things, we need to assign values to objects. To create an object, we need to give it a name followed by the assignment operator <-, and the value we want to give it:
weight_kg <- 55<- is the assignment operator. It assigns values on the right to objects on the left. So, after executing x <- 3, the value of x is 3. The arrow can be read as 3 goes into x. For historical reasons, you can also use = for assignments, but not in every context. Because of the slight differences in syntax, it is good practice to always use <- for assignments.
In RStudio, typing Alt + - (push Alt at the same time as the - key) will write <- in a single keystroke.
Objects can be given any name such as x, current_temperature, or subject_id. You want your object names to be explicit and not too long. They cannot start with a number (2x is not valid, but x2 is). R is case sensitive (e.g., weight_kg is different from Weight_kg). There are some names that cannot be used because they are the names of fundamental functions in R (e.g., if, else, for, see here for a complete list). In general, even if it’s allowed, it’s best to not use other function names (e.g., c, T, mean, data, df, weights). If in doubt, check the help to see if the name is already in use. It’s also best to avoid dots (.) within a variable name as in my.dataset. There are many functions in R with dots in their names for historical reasons, but because dots have a special meaning in R (for methods) and other programming languages, it’s best to avoid them. It is also recommended to use nouns for variable names, and verbs for function names. It’s important to be consistent in the styling of your code (where you put spaces, how you name variables, etc.). Using a consistent coding style makes your code clearer to read for your future self and your collaborators. In R, two popular style guides are Hadley Wickham’s and Google’s (this one is also comprehensive).
When assigning a value to an object, R does not print anything. You can force R to print the value by using parentheses or by typing the object name:
weight_kg <- 55 # doesn't print anything
(weight_kg <- 55) # but putting parenthesis around the call prints the value of `weight_kg`## [1] 55weight_kg # and so does typing the name of the object## [1] 55Now that R has weight_kg in memory, we can do arithmetic with it. For instance, we may want to convert this weight into pounds (weight in pounds is 2.2 times the weight in kg):
2.2 * weight_kg## [1] 121We can also change a variable’s value by assigning it a new one:
weight_kg <- 57.5
2.2 * weight_kg## [1] 126.5This means that assigning a value to one variable does not change the values of other variables. For example, let’s store the animal’s weight in pounds in a new variable, weight_lb:
weight_lb <- 2.2 * weight_kgand then change weight_kg to 100.
weight_kg <- 100What do you think is the current content of the object weight_lb? 126.5 or 220?
1.5.3 Functions and their arguments
Functions are “canned scripts” that automate more complicated sets of commands including operations assignments, etc. Many functions are predefined, or can be made available by importing R packages (more on that later). A function usually gets one or more inputs called arguments. Functions often (but not always) return a value. A typical example would be the function sqrt(). The input (the argument) must be a number, and the return value (in fact, the output) is the square root of that number. Executing a function (‘running it’) is called calling the function. An example of a function call is:
b <- sqrt(a)Here, the value of a is given to the sqrt() function, the sqrt() function calculates the square root, and returns the value which is then assigned to variable b. This function is very simple, because it takes just one argument.
The return ‘value’ of a function need not be numerical (like that of sqrt()), and it also does not need to be a single item: it can be a set of things, or even a dataset. We’ll see that when we read data files into R.
Arguments can be anything, not only numbers or filenames, but also other objects. Exactly what each argument means differs per function, and must be looked up in the documentation (see below). Some functions take arguments which may either be specified by the user, or, if left out, take on a default value: these are called options. Options are typically used to alter the way the function operates, such as whether it ignores ‘bad values’, or what symbol to use in a plot. However, if you want something specific, you can specify a value of your choice which will be used instead of the default.
Let’s try a function that can take multiple arguments: round().
round(3.14159)## [1] 3Here, we’ve called round() with just one argument, 3.14159, and it has returned the value 3. That’s because the default is to round to the nearest whole number. If we want more digits we can see how to do that by getting information about the round function. We can use args(round) or look at the help for this function using ?round.
args(round)## function (x, digits = 0)
## NULL?roundWe see that if we want a different number of digits, we can type digits=2 or however many we want.
round(3.14159, digits=2)## [1] 3.14If you provide the arguments in the exact same order as they are defined you don’t have to name them:
round(3.14159, 2)## [1] 3.14And if you do name the arguments, you can switch their order:
round(digits=2, x=3.14159)## [1] 3.14It’s good practice to put the non-optional arguments (like the number you’re rounding) first in your function call, and to specify the names of all optional arguments. If you don’t, someone reading your code might have to look up the definition of a function with unfamiliar arguments to understand what you’re doing.
1.5.4 Objects vs. variables
What are known as objects in R are known as variables in many other programming languages. Depending on the context, object and variable can have drastically different meanings. However, in this lesson, the two words are used synonymously. For more information see: https://cran.r-project.org/doc/manuals/r-release/R-lang.html#Objects
1.6 Vectors and data types
A vector is the most common and basic data type in R, and is pretty much the workhorse of R. A vector is composed by a series of values, which can be either numbers or characters. We can assign a series of values to a vector using the c() function. For example we can create a vector of peptide lengths and assign it to a new object pep_len:
pep_lens <- c(17, 13, 7)
pep_lens## [1] 17 13 7A vector can also contain characters:
peps <- c("VESITARHGEVLQLRPK", "IDGQWVTHQWLK", "LVILLFR")
peps## [1] "VESITARHGEVLQLRPK" "IDGQWVTHQWLK" "LVILLFR"The quotes around the peptide sequences are essential here. Without the quotes R will assume there are objects called VESITARHGEVLQLRPK, LVILLFR, … As these objects don’t exist in R’s memory, there will be an error message.
There are many functions that allow you to inspect the content of a vector. length() tells you how many elements are in a particular vector:
length(pep_lens)## [1] 3length(peps)## [1] 3An important feature of a vector, is that all of the elements are the same type of data. The function class() indicates the class (the type of element) of an object:
class(pep_lens)## [1] "numeric"class(peps)## [1] "character"The function str() provides an overview of the structure of an object and its elements. It is a useful function when working with large and complex objects:
str(pep_lens)## num [1:3] 17 13 7str(peps)## chr [1:3] "VESITARHGEVLQLRPK" "IDGQWVTHQWLK" "LVILLFR"You can use the c() function to add other elements to your vector:
pep_lens <- c(pep_lens, 5) # add to the end of the vector
pep_lens <- c(10, pep_lens) # add to the beginning of the vector
pep_lens## [1] 10 17 13 7 5In the first line, we take the original vector pep_lens, add the value 5 to the end of it, and save the result back into pep_lens. Then we add the value 10 to the beginning, again saving the result back into pep_lens.
We can do this over and over again to grow a vector, or assemble a dataset. As we program, this may be useful to add results that we are collecting or calculating.
We just saw 2 of the 6 main atomic vector types (or data types) that R uses: "character" and "numeric". These are the basic building blocks that all R objects are built from. The other 4 are:
"logical"forTRUEandFALSE(the boolean data type)"integer"for integer numbers (e.g.,2L, theLindicates to R that it’s an integer)"complex"to represent complex numbers with real and imaginary parts (e.g.,1+4i) and that’s all we’re going to say about them"raw"that we won’t discuss further
Vectors are one of the many data structures that R uses. Other important ones are lists (list), matrices (matrix), data frames (data.frame), factors (factor) and arrays (array).
Challenge
We’ve seen that atomic vectors can be of type character, numeric, integer, and logical. But what happens if we try to mix these types in a single vector?
What will happen in each of these examples? (hint: use
class()to check the data type of your objects):num_char <- c(1, 2, 3, 'a') num_logical <- c(1, 2, 3, TRUE) char_logical <- c('a', 'b', 'c', TRUE) tricky <- c(1, 2, 3, '4')
If we want to extract one or several values from a vector, we must provide one or several indices in square brackets. For instance:
peps <- c("VESITARHGEVLQLRPK", "IDGQWVTHQWLK", "LVILLFR", "ARHGILPK")
peps[2]## [1] "IDGQWVTHQWLK"peps[c(3, 2)]## [1] "LVILLFR" "IDGQWVTHQWLK"We can also repeat the indices to create an object with more elements than the original one:
more_peps <- peps[c(1, 2, 3, 2, 1, 4)]
more_peps## [1] "VESITARHGEVLQLRPK" "IDGQWVTHQWLK" "LVILLFR"
## [4] "IDGQWVTHQWLK" "VESITARHGEVLQLRPK" "ARHGILPK"R indices start at 1. Programming languages like Fortran, MATLAB, and R start counting at 1, because that’s what human beings typically do. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that’s simpler for computers to do.
1.6.1 Conditional subsetting
Another common way of subsetting is by using a logical vector. TRUE will select the element with the same index, while FALSE will not:
pep_lens <- c(17, 12, 7, 8)
pep_lens[c(TRUE, FALSE, TRUE, FALSE)]## [1] 17 7Typically, these logical vectors are not typed by hand, but are the output of other functions or logical tests. For instance, if you wanted to select only the values above 50:
pep_lens > 10 # will return logicals with TRUE for the indices that meet the condition## [1] TRUE TRUE FALSE FALSE## so we can use this to select only the values above 10
pep_lens[pep_lens > 10]## [1] 17 12You can combine multiple tests using & (both conditions are true, AND) or | (at least one of the conditions is true, OR):
pep_lens[pep_lens < 8 | pep_lens > 15]## [1] 17 7pep_lens[pep_lens <= 12 & pep_lens == 10]## numeric(0)Here, < stands for “less than”, > for “greater than”, >= for “greater than or equal to”, and == for “equal to”. The double equal sign == is a test for numerical equality between the left and right hand sides, and should not be confused with the single = sign, which performs variable assignment (similar to <-).
A common task is to search for certain strings in a vector. One could use the “or” operator | to test for equality to multiple values, but this can quickly become tedious. The function %in% allows you to test if any of the elements of a search vector are found:
peps <- c("VESITARHGEVLQLRPK", "IDGQWVTHQWLK", "LVILLFR", "ARHGILPK")
peps[peps == "IDGQWVTHQWLK" | peps == "LVILLFR"] # returns both peptides## [1] "IDGQWVTHQWLK" "LVILLFR"peps %in% c("IDGQWVTHQWLK", "LVILLFR", "SITARH", "VESITA", "ARHGILGHIIHKKP")## [1] FALSE TRUE TRUE FALSEpeps[peps %in% c("IDGQWVTHQWLK", "LVILLFR", "SITARH", "VESITA", "ARHGILGHIIHKKP")]## [1] "IDGQWVTHQWLK" "LVILLFR"1.7 Missing data
As R was designed to analyze datasets, it includes the concept of missing data (which is uncommon in other programming languages). Missing data are represented in vectors as NA.
When doing operations on numbers, most functions will return NA if the data you are working with include missing values. This feature makes it harder to overlook the cases where you are dealing with missing data. You can add the argument na.rm=TRUE to calculate the result while ignoring the missing values.
heights <- c(2, 4, 4, NA, 6)
mean(heights)## [1] NAmax(heights)## [1] NAmean(heights, na.rm = TRUE)## [1] 4max(heights, na.rm = TRUE)## [1] 6If your data include missing values, you may want to become familiar with the functions is.na(), na.omit(), and complete.cases(). See below for examples.
## Extract those elements which are not missing values.
heights[!is.na(heights)]## [1] 2 4 4 6## Returns the object with incomplete cases removed. The returned object is atomic.
na.omit(heights)## [1] 2 4 4 6
## attr(,"na.action")
## [1] 4
## attr(,"class")
## [1] "omit"## Extract those elements which are complete cases.
heights[complete.cases(heights)]## [1] 2 4 4 6Challenge
Using this vector of length measurements, create a new vector with the NAs removed.
lens <- c(10, 24, NA, 18, NA, 20)Use the function
median()to calculate the median of thelensvector.
lens <- c(10, 24, NA, 18, NA, 20)
median(lens) ## NA## [1] NAlens2 <- na.omit(lens)
median(lens)## [1] NAmedian(lens, na.rm = TRUE)## [1] 19Now that we have learned how to write scripts, and the basics of R’s data structures, we are ready to start working with the Portal dataset we have been using in the other lessons, and learn about data frames.
1.8 R markdown
Markdown (extension .md) is a very simple markup language in plain text, that can be converted to many different outputs, such as pdf and html to name a few.
It is also possible to interleave R code into markdown documents, producing so called R mardown documents (extension .Rmd). Before generating the final html of pdf output, the Rmd files are first knit (or weaved), i.e. the R code chunks are extracted, exectuted and replaced by their output, be it text, tables or figures.
Rmarkdown is a fantastic tool to implement reproducible research, as we have the guarantee that code results and figures, displayed in the final document, accurately represent the analysis restults. They provide a relatively easly way to generate high quality and reproducible analysis reports for any arbitrary input data that is submitted to an established analysis routine. The R package that supports this is rmarkdown, and the whole process is made relatively easy with RStudio and available documentation.
We are going to walk through the creation of such a document.
Challenge
- Create a new R markdown document in RStudio, and compile it to html or pdf.
- An important aspect of reproducible research is the record the version of software used. Add a code chunk at the end of the R markdown document and call
sessionInfo.
In the next sections, we are going to learn how to read data sets into R, how to explore and visualise the data, and how to perform basic statistical analyses.
1.5.2 Comments
The comment character in R is
#, anything to the right of a#in a script will be ignored by R. It is useful to leave notes, and explanations in your scripts. RStudio makes it easy to comment or uncomment a paragraph: after selecting the lines you want to comment, press at the same time on your keyboard Ctrl + Shift + C.