MSnbase benchmarking
Laurent Gatto
de Duve Institute, UCLouvain, BelgiumJohannes Rainer
Center for Biomedicine, EURAC, Bolzano, ItalySource:
vignettes/v04-benchmarking.Rmd
v04-benchmarking.RmdIntroduction
In this vignette, we will document various timings and benchmarkings of the MSnbase version 2, that focuses on on-disk data access (as opposed to in-memory). More details about the new implementation are documented in the respective classes manual pages and in
MSnbase, efficient and elegant R-based processing and visualisation of raw mass spectrometry data. Laurent Gatto, Sebastian Gibb, Johannes Rainer. bioRxiv 2020.04.29.067868; doi: https://doi.org/10.1101/2020.04.29.067868
As a benchmarking dataset, we are going to use a subset of an TMT 6-plex experiment acquired on an LTQ Orbitrap Velos, that is distributed with the msdata package
library("msdata")
f <- msdata::proteomics(full.names = TRUE,
pattern = "TMT_Erwinia_1uLSike_Top10HCD_isol2_45stepped_60min_01.mzML.gz")
basename(f)## [1] "TMT_Erwinia_1uLSike_Top10HCD_isol2_45stepped_60min_01.mzML.gz"We need to load the MSnbase
package and set the session-wide verbosity flag to
FALSE.
library("MSnbase")
setMSnbaseVerbose(FALSE)Benchmarking
Reading data
We first read the data using the original behaviour
readMSData function by setting the mode
argument to "inMemory" to generates an in-memory
representation of the MS2-level raw data and measure the time needed for
this operation.
system.time(inmem <- readMSData(f, msLevel = 2,
mode = "inMemory",
centroided = TRUE))## user system elapsed
## 3.957 0.097 4.046Next, we use the readMSData function to generate an
on-disk representation of the same data by setting
mode = "onDisk".
system.time(ondisk <- readMSData(f, msLevel = 2,
mode = "onDisk",
centroided = TRUE))## user system elapsed
## 1.320 0.031 1.344Creating the on-disk experiment is considerable faster and scales to much bigger, multi-file data, both in terms of object creation time, but also in terms of object size (see next section). We must of course make sure that these two datasets are equivalent:
all.equal(inmem, ondisk)## [1] TRUEData size
To compare the size occupied in memory of these two objects, we are
going to use the object_size function from the pryr package,
which accounts for the data (the spectra) in the assayData
environment (as opposed to the object.size function from
the utils package).
library("pryr")
object_size(inmem)## 2.77 MB
object_size(ondisk)## 242.01 kBThe difference is explained by the fact that for ondisk,
the spectra are not created and stored in memory; they are access on
disk when needed, such as for example for plotting:
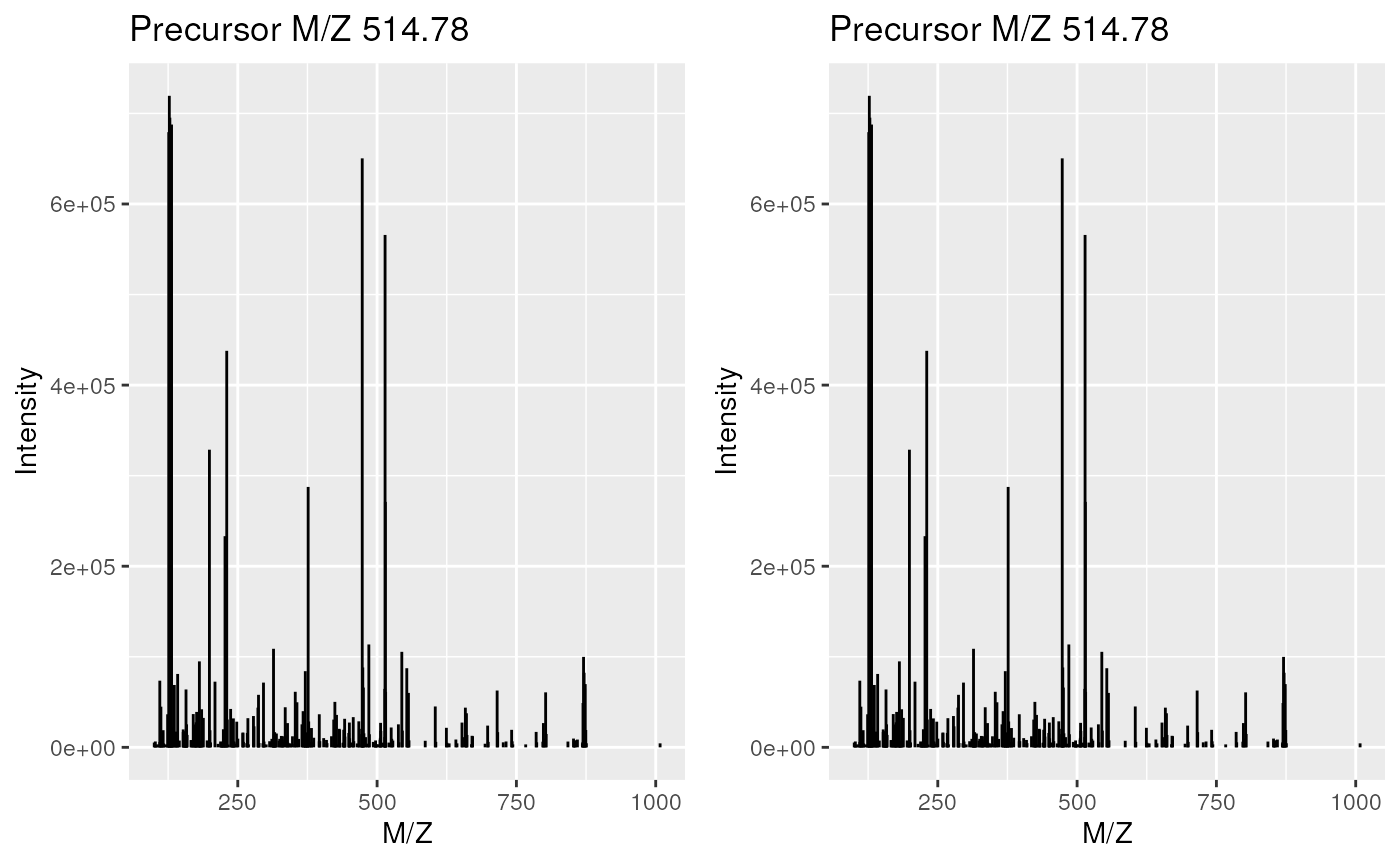
Plotting in-memory and on-disk spectra
Accessing spectra
The drawback of the on-disk representation is when the spectrum data has to actually be accessed. To compare access time, we are going to use the microbenchmark and repeat access 10 times to compare access to all 451 and a single spectrum in-memory (i.e. pre-loaded and constructed) and on-disk (i.e. on-the-fly access).
library("microbenchmark")
mb <- microbenchmark(spectra(inmem),
inmem[[200]],
spectra(ondisk),
ondisk[[200]],
times = 10)
mb## Unit: microseconds
## expr min lq mean median uq
## spectra(inmem) 67.076 78.937 156.1754 178.9140 194.773
## inmem[[200]] 16.832 19.546 43.1573 45.3845 63.999
## spectra(ondisk) 292989.921 294197.756 296390.1532 294300.6380 297409.269
## ondisk[[200]] 163645.230 165293.236 167278.4735 166761.0850 168249.793
## max neval
## 236.822 10
## 70.722 10
## 309262.368 10
## 174095.642 10While it takes order or magnitudes more time to access the data on-the-fly rather than a pre-generated spectrum, accessing all spectra is only marginally slower than accessing all spectra, as most of the time is spent preparing the file for access, which is done only once.
On-disk access performance will depend on the read throughput of the
disk. A comparison of the data import of the above file from an internal
solid state drive and from an USB3 connected hard disk showed only small
differences for the onDisk mode (1.07 vs 1.36
seconds), while no difference were observed for accessing individual or
all spectra. Thus, for this particular setup, performance was about the
same for SSD and HDD. This might however not apply to setting in which
data import is performed in parallel from multiple files.
Data access does not prohibit interactive usage, such as plotting, for example, as it is about 1/2 seconds, which is an operation that is relatively rare, compared to subsetting and filtering, which are faster for on-disk data:
i <- sample(length(inmem), 100)
system.time(inmem[i])## user system elapsed
## 0.077 0.000 0.077
system.time(ondisk[i])## user system elapsed
## 0.011 0.000 0.011Operations on the spectra data, such as peak picking, smoothing, cleaning, … are cleverly cached and only applied when the data is accessed, to minimise file access overhead. Finally, specific operations such as for example quantitation (see next section) are optimised for speed.
MS2 quantitation
Below, we perform TMT 6-plex reporter ions quantitation on the first 100 spectra and verify that the results are identical (ignoring feature names).
system.time(eim <- quantify(inmem[1:100], reporters = TMT6,
method = "max"))## user system elapsed
## 6.288 1.288 4.060
system.time(eod <- quantify(ondisk[1:100], reporters = TMT6,
method = "max"))## user system elapsed
## 0.222 0.081 0.272
all.equal(eim, eod, check.attributes = FALSE)## [1] TRUENotable differences on-disk and in-memory implementations
The MSnExp and OnDiskMSnExp documentation
files and the MSnbase developement vignette provide more
information about implementation details.
MS levels
On-disk support multiple MS levels in one object, while in-memory only supports a single level. While support for multiple MS levels could be added to the in-memory back-end, memory constrains make this pretty-much useless and will most likely never happen.
Serialisation
In-memory objects can be save()ed and
load()ed, while on-disk can’t. As a workaround,
the latter can be coerced to in-memory instances with
as(, "MSnExp"). We would need mzML write
support in mzR to be
able to implement serialisation for on-disk data.
Conclusions
This document focuses on speed and size improvements of the new
on-disk MSnExp representation. The extend of these
improvements will substantially increase for larger data.
For general functionality about the on-disk MSnExp data
class and MSnbase
in general, see other vignettes available with
vignette(package = "MSnbase")