CCP LOPIT hackathon
Introduction
Today (14 July 2015), we had our first CCP LOPIT hackathon. LOPIT [1,2] is a mass spectrometry-based technique to systematically map thousands of proteins to their sub-cellular niches, typically organelles, but also macro-molecular complexes. This is very relevant from a biological point of view, as proteins can only function and/or interact with their partners if they effectively make it to their expected destination. Abnormal protein localisation has been show to lead to the loss of functional effects in diseases (Laurila and Vihinen, 2009); another example is disruption of the nuclear/cytoplasmic transport (nuclear pores) that have been detected in many types of carcinoma cells (Kau et al., 2004). For a general introduction on LOPIT and spatial proteomics, see this review.
I have been working on the computational side of spatial proteomics since 2010, and have described the current state of the art here.

Material and methods
The hackathon brought together a dozen members of the Cambridge Centre for Proteomics to mine a recent mouse embryonic stem cell data (currently unpublished). Most people were biologists that knew the LOPIT technique and these kind of data quite well. I generated 300+ precompiled protein sets of interest (all KEGG pathways, and user contributed protein sets) and mapped them to the data, generated template reports and figures highlighting the major sub-cellular niches and the protein sets of interest, and set up a simple GUI to help the participants navigate the data and search for other proteins of interest. All these were centralised in a github repository.
Results
The figure below illustrates some observed proteins involved in
protein export
(KEGG pathway mmu03060)
on the LOPIT PCA plot, where coloured clusters represent well-known
sub-cellular niches.
The protein export is the active transport of proteins from the cytoplasm to the exterior of the cell, or to the periplasmic compartment in Gram-negative bacteria.
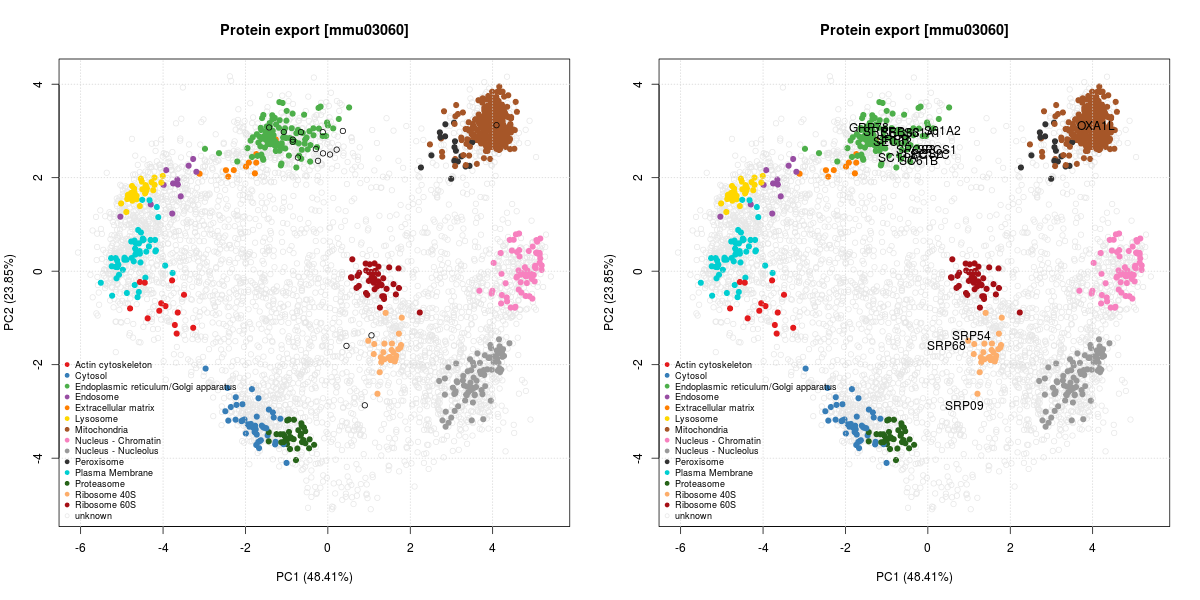
We can see that many proteins localise in the ER cluster. Others, like the Signal recognition particle 9 kDa protein (SRP9) is documented as being cytoplasmic with a endoplasmic reticulum targeting (which diffuses to the endoplasmic reticulum):
Signal-recognition-particle assembly has a crucial role in targeting secretory proteins to the rough endoplasmic reticulum membrane. SRP9 together with SRP14 (which we haven’t observed in our data) and the Alu portion of the SRP RNA, constitutes the elongation arrest domain of SRP. The complex of SRP9 and SRP14 is required for SRP RNA binding.
This is compatible with its localisation on the PCA plots, between the
cytosol (blue) and the ER (green). SRP54 and SRP68 show similar,
compatible patterns.
Now imagine what 10+ competent biologists, that know the assays and the type of data, know a lot about proteins, their biochemistry and biochemical pathways, armed with the right tools, can do in a day of focused work!
Conclusions and perspectives
The result has been great. We gathered 20+ annotated protein sets, some in great details by people that have substantial expertise. There are great opportunities for such events, with biologists, bioinformaticians, and a simple, but dedicated infrastructure to prioritise our efforts on the interpretation of data.
One thing that will need some thoughts is an effective and easy way for reporting. I generated templates and figures for each protein set of interest, but even with the gihub editor, this is not enough. Github issues enable to easily upload or drag and drop figures, but this is not available for regular markdown files. I did not have time to generate a github issue per protein set, containing all the default descriptions and links, but this should be possible.

