May Institue keynote talk: Reproducible mass spectrometry-based research
These are my notes for my talk at the 2021 edition of the May Institute in Computation and statistics for mass spectrometry and proteomics.
Acknowledgement
I would like to start by thanking the May Institute organisers for the invitation, and for organising such an event. The reasons why I think their efforts are so valuable should become apparent below. For now, suffice to say that I believe that it is essential for researchers to be able to question their data without constrains - and we need the tools and the skills to be able to do that. The May Institute is one place where we can acquire or improve these skills.
Motivation
Inverse problems are hard!
Example and figure borrowed from Stephen Eglen.
| Score (%) | grade |
|---|---|
| 70-100 | A |
| 60-69 | B |
| 50-59 | C |
| 40-49 | D |
| 0-39 | F |
-
Forward problem: I scored 68, what was my grade?
-
Inverse problem: I got a B, what was my score?
Research sharing: the inverse problem
Where is the scholarship?
An article about computational science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and that complete set of instructions that generated the figures.
[Buckheit and Donoho 1995, after Claerbout]
Why reproducibility is important
For scientific reasons: think reproducibility crisis (discussed below).
For political reasons: public trust in science, in data, in experts; without (public) trust in science and research, there won’t be any funding anymore; lack of (public) trust in science leads to poor public health decisions!
Reliable research
It’s not about reprodcible research.
It’s not about open research.
It’s about reliable research! It’s about trust – openness and reproducibility are means that (should) lead to greater trust in the data, analyses and results.
Is there a cost?
Well, yes, probably … there’s no such thing as a free lunch.
But
- the cost is worth it IMHO
- cost is mostly time, (in most cases) not $.
- with the right tools and skills, these costs can be minimised
- and it requires discipline.
Actually no, it is a matter or relocating time!
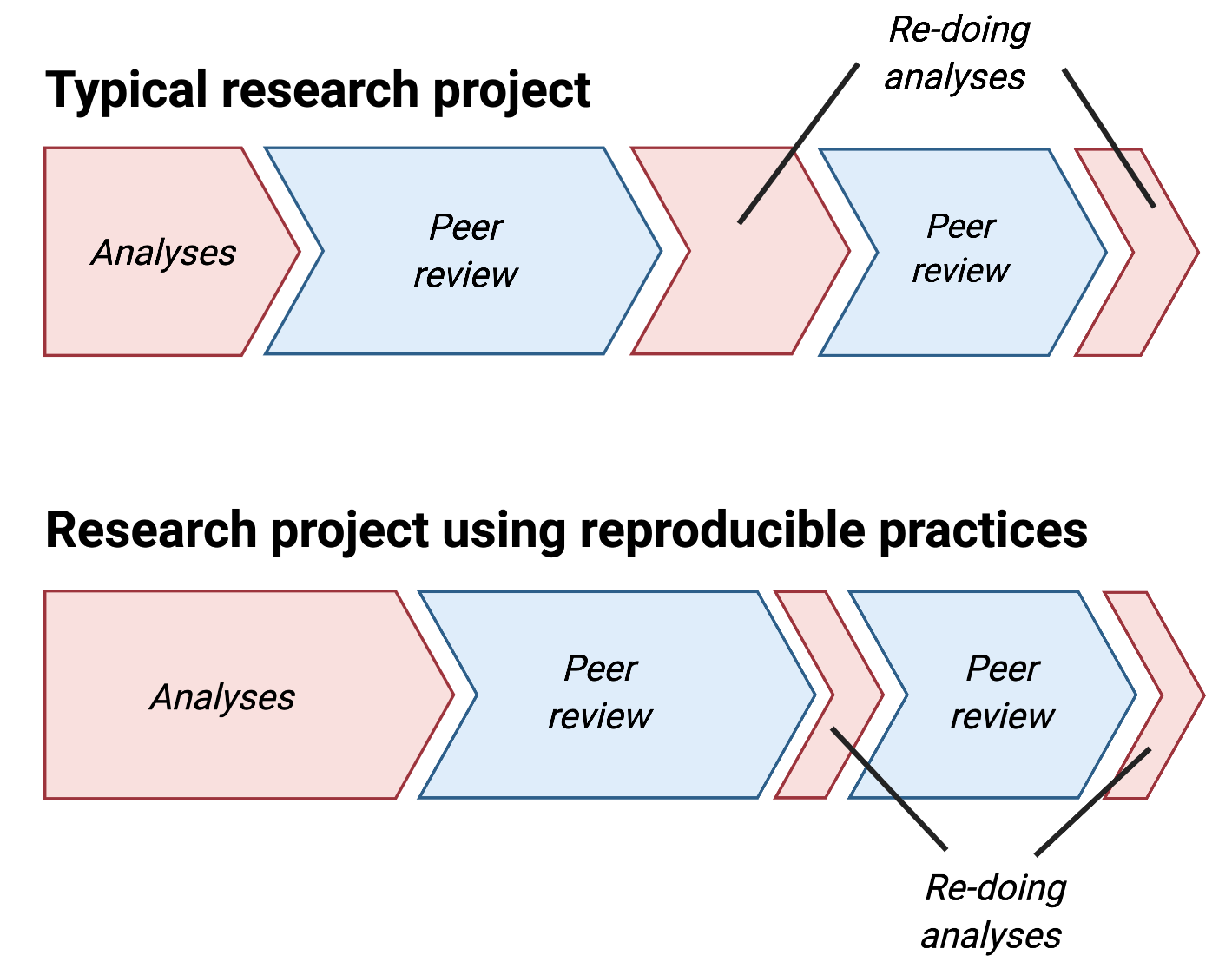
From Five things about open and reproducible science that every early career researcher should know.
But what do we mean by reproducibility?
From a But what to we mean by reproducibility? blog post.
-
Repeat my experiment, i.e. obtain the same tables/graphs/results using the same setup (data, software, …) in the same lab or on the same computer. That’s basically re-running one of my analysis some time after I original developed it.
-
Reproduce an experiment (not mine), i.e. obtain the same tables/graphs/results in a different lab or on a different computer, using the same setup (the data would be downloaded from a public repository and the same software, but possibly different version, different OS, is used). I suppose, we should differentiate replication using a fresh install and a virtual machine or docker image that replicates the original setup.
-
Replicate an experiment, i.e. obtain the same (similar enough) tables/graphs/results in a different set up. The data could still be downloaded from the public repository, or possibly re-generate/re-simulate it, and the analysis would be re-implemented based on the original description. This requires openness, and one would clearly not be allowed the use a black box approach (VM, docker image) or just re-running a script.
-
Finally, re-use the information/knowledge from one experiment to run a different experiment with the aim to confirm results from scratch.
Another view (from a talk by Kirstie Whitaker):
| Same Data | Different Data | |
|---|---|---|
| Same Code | reproduce | replicate |
| Different Code | robust | generalisable |
See also this opinion piece by Jeffrey T. Leek and Roger D. Peng, Reproducible research can still be wrong: Adopting a prevention approach.
Whether you want to
- repeat the analysis you just did yesterday;
- reproduce some old code on the same or different data;
- improve your code using existing data;
- replicate someone else’s analysis using their data (but your code/software); or
- generate new data to verify/validate the scientific validity/replicability of a study
in my opinion, it all boils down to understanding what has been done and what is happening.
I am not (that much) interested in blindly re-running something to confirm that I can the same results without understanding it. However, it is important to acknowledge that even simply repeating or reproducing a complex analysis isn’t trivial - especially for very large data, large compute requirement, dedicate hardware, long simulation times, …
Note that sometimes, things are expected not the reproduce, for example when bugs are fixed, analysis methods are changed/improved. This brings us back to the importance of understanding what we do and why we do it.
Why reproducibility is important (as an individual researcher)
From
Gabriel Becker An Imperfect Guide to Imperfect Reproducibility, May Institute for Computational Proteomics, 2019.
(Computational) Reproducibility Is Not The Point
Take home message:
The goal is trust, verification and guarantees:
- Trust in Reporting - result is accurately reported
- Trust in Implementation - analysis code successfully implements chosen methods
- Statistical Trust - data and methods are (still) appropriate
- Scientific Trust - result convincingly supports claim(s) about underlying systems or truths
Reproducibility As A Trust Scale (copyright Genentech Inc)
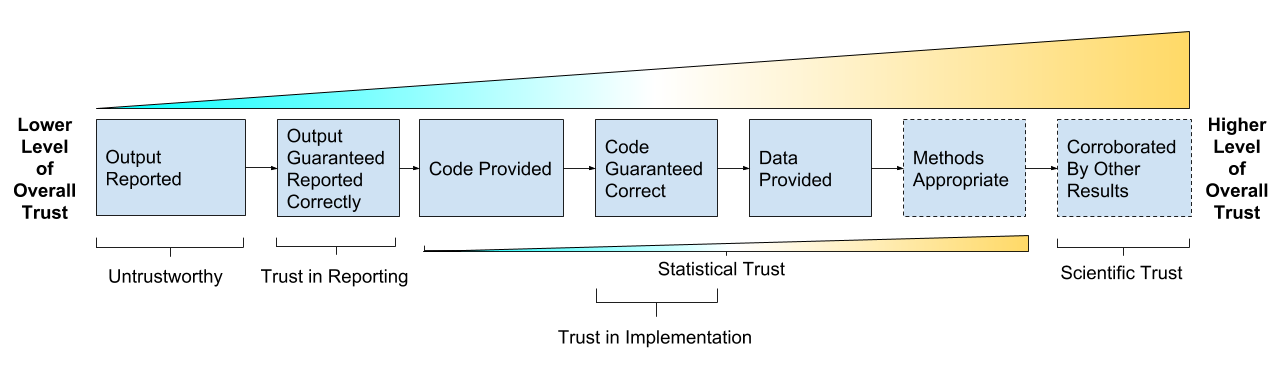
Take home message:
Reproducibility isn’t binary, it’s a gradient, it’s multidisciplinary, it’s multidimensional.
Another take home message:
Reproducibility isn’t easy.
More reasons to become a reproducible research practitioner
Florian Markowetz, Five selfish reasons to work reproducibly, Genome Biology 2015, 16:274.
And so, my fellow scientists: ask not what you can do for reproducibility; ask what reproducibility can do for you! Here, I present five reasons why working reproducibly pays off in the long run and is in the self-interest of every ambitious, career-oriented scientist.
- Reproducibility helps to avoid disaster: a project is more than a beautiful result. You need to record in detail how you got there. Starting to work reproducibly early on will save you time later. I had cases where a collaborator told me they preferred the results on the very first plots they received, that I couldn’t recover a couple of month later. But because my work was reproducible and I had tracked it over time (using git and GitHub), I was able, after a little bit of data analysis forensics, to identify why these first, preliminary plots weren’t consistent with later results (and it as a simple, but very relevant bug in the code). Imagine if my collaborators had just used these first plots for publication, or to decide to perform further experiments.
- Reproducibility makes it easier to write papers: Transparency in your analysis makes writing papers much easier. In dynamic documents (using rmarkdown, juypter notebook and other similar tools), all results are automatically update when the data are changed. You can be confident your numbers, figures and tables are up-to-date.
- Reproducibility helps reviewers see it your way: a reproducible document will tick many of the boxes enumerated above. You will make me very happy reviewer if I can review a paper that is reproducible.
- Reproducibility enables continuity of your work: quoting Florian, “In my own group, I don’t even discuss results with students if they are not documented well. No proof of reproducibility, no result!”.
- Reproducibility helps to build your reputation: publishing reproducible research will build you the reputation of being an honest and careful researcher. In addition, should there ever be a problem with a paper, a reproducible analysis will allow to track the error and show that you reported everything in good faith.
Tools for reproducible research
- R/Bioconductor – many of you are familiar or even experts with R.
- Writing R scripts (could also be Rmd files) to keep track of what we do, and be able to easily re-run the code.
Here (i.e. as part of the May Institute), I took the decision to let you install the packages on your computers. Many of you will have taken the R classes, so you should have a working installation and good to go.
Another approach could have been to prepare Docker container or Amazon Machine Image with all pre-installed software and data (Bioconductor provides Docker containers and AMIs). Bioconductor uses the Orchestra platform for conference workshops. I use the Renku environment for my university teaching. After installing docker on your computer or running an pre-configured online environment, you would simply download and run that container. This is particularly relevant for complex installations.
In my experience, the installation of R and R/Bioconductor packages is relatively straighforward (and a useful experience) to be done by hand by researchers that want to analyse their own data.
- Packages from the R for Mass Spectrometry initiative
The R for Mass Spectrometry initiative

The aim of the R for Mass Spectrometry initiative is to provide efficient, thoroughly documented, tested and flexible R software for the analysis and interpretation of high throughput mass spectrometry assays, including proteomics and metabolomics experiments. The project formalises the longtime collaborative development efforts of its core members under the R for Mass Spectrometry organisation to facilitate dissemination and accessibility of their work.
- Laurent Gatto, UCLouvain, Belgium
- Sebastian Gibb, University Medicine Greifswald, Germany
- Johannes Rainer, Eurac Research, Italy.
More info at
- Project page: https://www.rformassspectrometry.org/
- Individual package pages
- Tutorial: https://rformassspectrometry.github.io/docs/
Live coding
I’ll focus on raw data infrastructure, centred around the Spectra package. For quantitative data, see the QFeatures package.
- Package installation
- Downloading data
- Make an experiment, adding files, possible other metadata (pmid, doi)
- create raw data object
- load identification data
- filter id data
- join spectra and id data
- Add them to the experiment
- Optionally also add id data
- Extract subset of interest
- Change backend
- Cluster spectra
- Visualise data
The code is available here.



