Storing multiple ClustDist instances
ClustDistList-class.RdA class for storing lists of ClustDist
instances.
Slots
x:Object of class
listcontaining validClustDistinstances.log:Object of class
listcontaining an object creation log, containing among other elements the call that generated the object..__classVersion__:The version of the instance. For development purposes only.
Methods
"[["Extracts a single
ClustDistat position."["Extracts one of more
ClustDistsasClustDistList.lengthReturns the number of
ClustDists.namesReturns the names of
ClustDists, if available. The replacement method is also available.showDisplay the object by printing a short summary.
lapply(x, FUN, ...)Apply function
FUNto each element of the inputx. If the application ofFUNreturns andClustDist, then the return value is anClustDistList, otherwise alist
.
plotPlots a boxplot of the distance results per protein set.
Examples
library('pRolocdata')
data(dunkley2006)
par <- setAnnotationParams(inputs =
c("Arabidopsis thaliana genes",
"Gene stable ID"))
#> Using species Arabidopsis thaliana genes (TAIR10)
#> Warning: Ensembl will soon enforce the use of https.
#> Ensure the 'host' argument includes "https://"
#> Using feature type Gene stable ID(s) [e.g. AT1G01010]
#> Connecting to Biomart...
#> Warning: Ensembl will soon enforce the use of https.
#> Ensure the 'host' argument includes "https://"
## add protein set/annotation information
xx <- addGoAnnotations(dunkley2006, par)
## filter
xx <- filterMinMarkers(xx, n = 50)
#> Retaining 3 out of 79 in GOAnnotations
xx <- filterMaxMarkers(xx, p = .25)
#> Retaining 2 out of 3 in GOAnnotations
## get distances for protein sets
dd <- clustDist(xx)
#>
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
## plot distances for all protein sets
plot(dd)
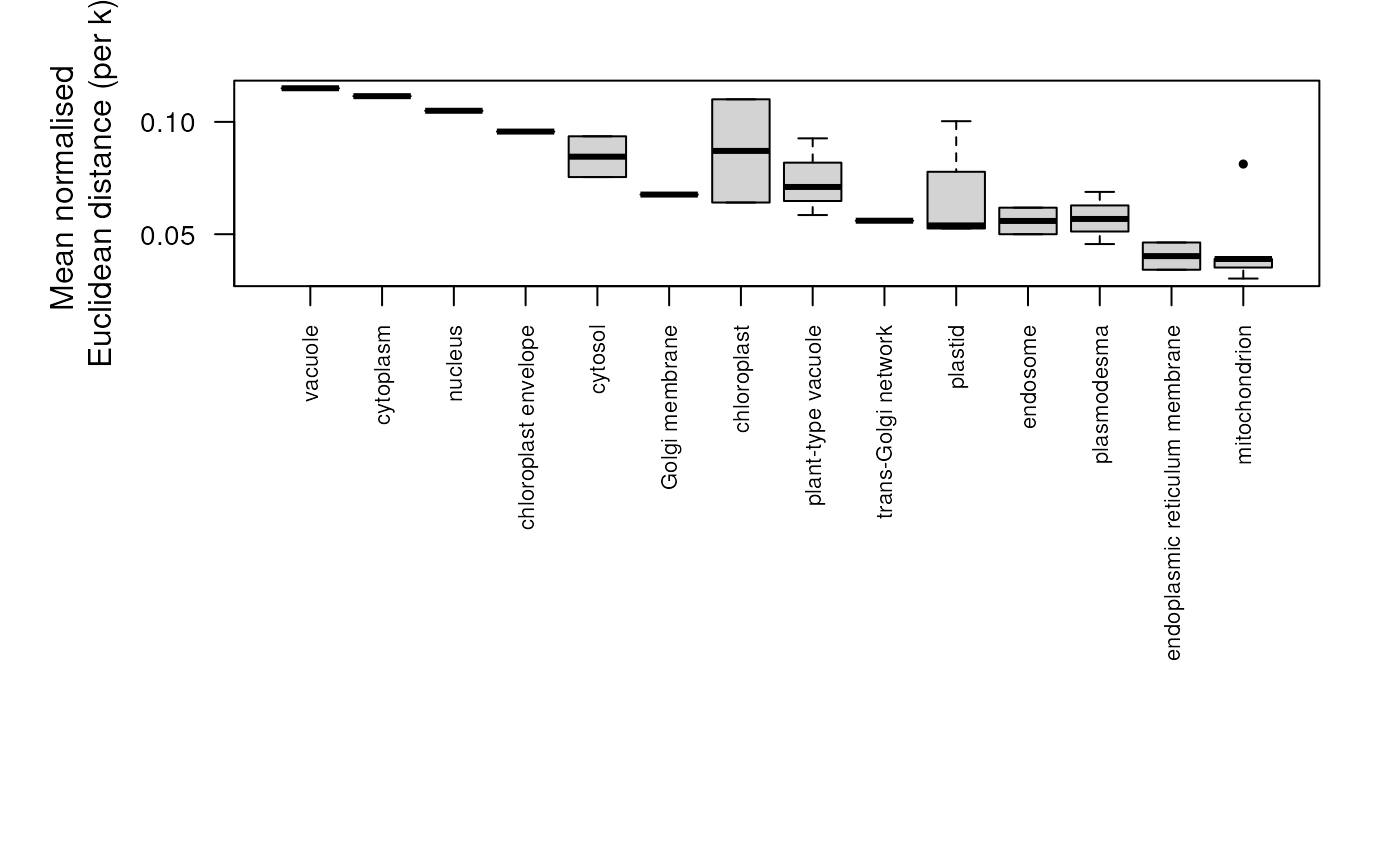 names(dd)
#> [1] "endoplasmic reticulum" "Golgi apparatus"
## Extract a sub-list of ClustDist objects
dd[1]
#> Instance of class 'ClustDistList' containig 1 objects.
## Extract 1st ClustDist object
dd[[1]]
#> Object of class "ClustDist"
#> fcol = GOAnnotations
#> term = GO:0005783
#> id = endoplasmic reticulum
#> nrow = 91
#> k's tested: 1 2 3 4 5
#> Size: 91
#> Size: 88
#> Size: 80
#> Size: NA
#> Size: NA
#> Clusters info:
#> ks.mean mean ks.norm norm
#> k = 1 1 0.1916 1 0.04260
#> k = 2 1 0.1595 1 0.03585
#> k = 3 1 *0.1385 1 *0.03215
#> k = 4 NA NA NA NA
#> k = 5 NA NA NA NA
names(dd)
#> [1] "endoplasmic reticulum" "Golgi apparatus"
## Extract a sub-list of ClustDist objects
dd[1]
#> Instance of class 'ClustDistList' containig 1 objects.
## Extract 1st ClustDist object
dd[[1]]
#> Object of class "ClustDist"
#> fcol = GOAnnotations
#> term = GO:0005783
#> id = endoplasmic reticulum
#> nrow = 91
#> k's tested: 1 2 3 4 5
#> Size: 91
#> Size: 88
#> Size: 80
#> Size: NA
#> Size: NA
#> Clusters info:
#> ks.mean mean ks.norm norm
#> k = 1 1 0.1916 1 0.04260
#> k = 2 1 0.1595 1 0.03585
#> k = 3 1 *0.1385 1 *0.03215
#> k = 4 NA NA NA NA
#> k = 5 NA NA NA NA