knn transfer learning classification
Source:R/machinelearning-functions-knntl.R
knntlClassification.RdClassification using a variation of the KNN implementation of Wu and Dietterich's transfer learning schema
Usage
knntlClassification(
primary,
auxiliary,
fcol = "markers",
bestTheta,
k,
scores = c("prediction", "all", "none"),
seed
)Arguments
- primary
An instance of class
"MSnSet".- auxiliary
An instance of class
"MSnSet".- fcol
The feature meta-data containing marker definitions. Default is
markers.- bestTheta
Best theta vector as output from
knntlOptimisation, seeknntlOptimisationfor details- k
Numeric vector of length 2, containing the best
kparameters to use for the primary and auxiliary datasets. If kkis not specified it will be calculated internally.- scores
One of
"prediction","all"or"none"to report the score for the predicted class only, for all classes or none.- seed
The optional random number generator seed.
Examples
# \donttest{
library(pRolocdata)
data(andy2011)
data(andy2011goCC)
## reducing calculation time of k by pre-running knnOptimisation
x <- c(andy2011, andy2011goCC)
k <- lapply(x, function(z)
knnOptimisation(z, times=5,
fcol = "markers.orig",
verbose = FALSE))
k <- sapply(k, function(z) getParams(z))
k
#> k k
#> 7 3
## reducing parameter search with theta = 1,
## weights of only 1 or 0 will be considered
opt <- knntlOptimisation(andy2011, andy2011goCC,
fcol = "markers.orig",
times = 2,
by = 1, k = k)
#> Removing 389 columns with only 0s.
#> Weigths:
#> (0, 1)
opt
#> Object of class "ThetaRegRes"
#> Algorithm: theta
#> Theta hyper-parameters:
#> weights: 0 1
#> k: 7 3
#> nrow: 16
#> Design:
#> Replication: 2 x 5-fold X-validation
#> Partitioning: 0.2/0.8 (test/train)
#> Results
#> macro F1:
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> 0.8327 0.8442 0.8558 0.8558 0.8673 0.8789
#> best theta:
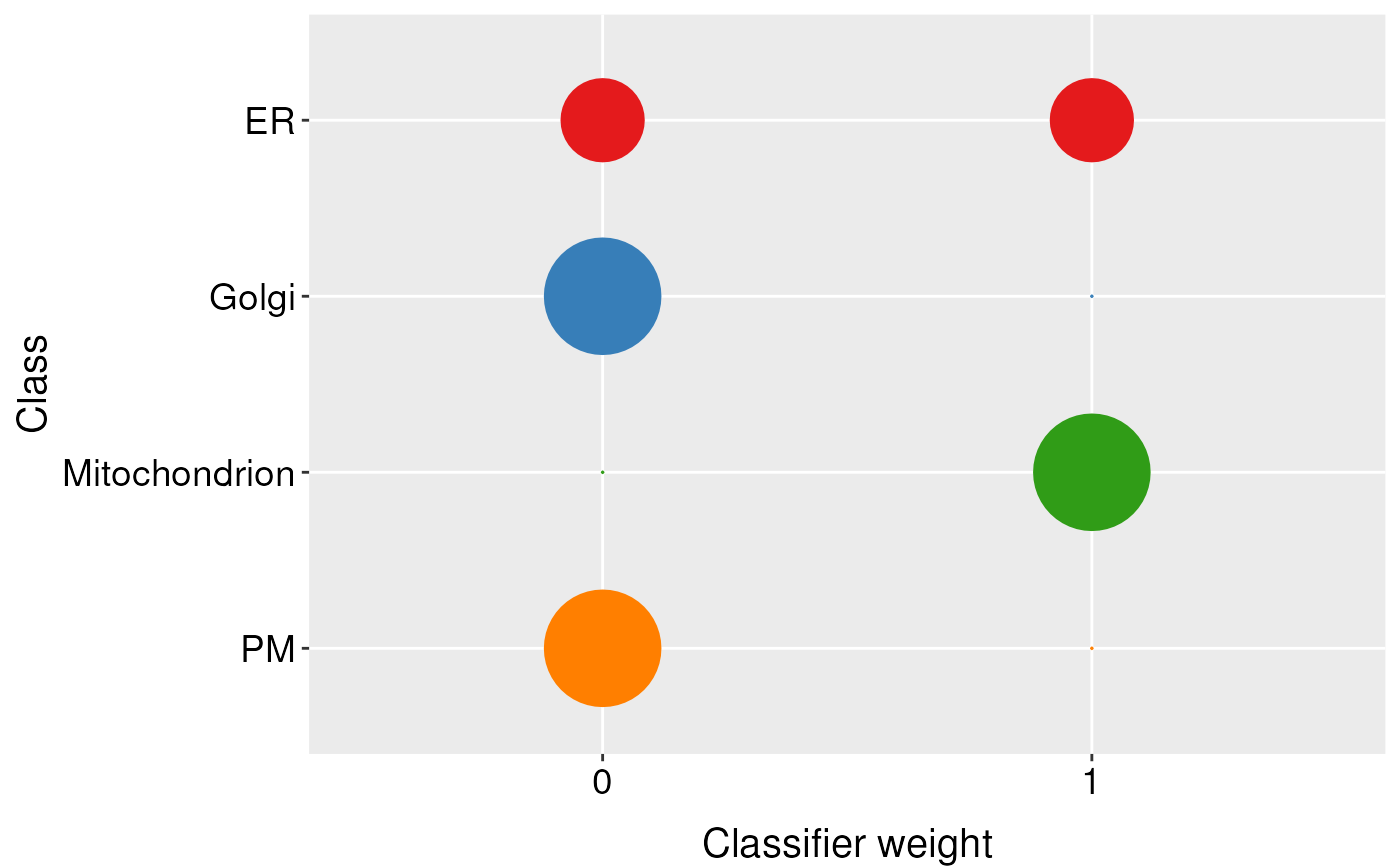
#> ER Golgi Mitochondrion PM
#> weight:0 1 2 0 1
#> weight:1 1 0 2 1
th <- getParams(opt)
plot(opt)
 res <- knntlClassification(andy2011, andy2011goCC,
fcol = "markers.orig", th, k)
res
#> MSnSet (storageMode: lockedEnvironment)
#> assayData: 1371 features, 8 samples
#> element names: exprs
#> protocolData: none
#> phenoData
#> sampleNames: X113 X114 ... X121 (8 total)
#> varLabels: Fraction.information
#> varMetadata: labelDescription
#> featureData
#> featureNames: O00767 P51648 ... O75312 (1371 total)
#> fvarLabels: Accession.No. Protein.Description ... knntl (12 total)
#> fvarMetadata: labelDescription
#> experimentData: use 'experimentData(object)'
#> Annotation:
#> - - - Processing information - - -
#> Loaded on Fri Sep 23 15:43:47 2016.
#> Normalised to sum of intensities.
#> Added markers from 'mrk' marker vector. Fri Sep 23 15:43:47 2016
#> MSnbase version: 1.99.2
# }
res <- knntlClassification(andy2011, andy2011goCC,
fcol = "markers.orig", th, k)
res
#> MSnSet (storageMode: lockedEnvironment)
#> assayData: 1371 features, 8 samples
#> element names: exprs
#> protocolData: none
#> phenoData
#> sampleNames: X113 X114 ... X121 (8 total)
#> varLabels: Fraction.information
#> varMetadata: labelDescription
#> featureData
#> featureNames: O00767 P51648 ... O75312 (1371 total)
#> fvarLabels: Accession.No. Protein.Description ... knntl (12 total)
#> fvarMetadata: labelDescription
#> experimentData: use 'experimentData(object)'
#> Annotation:
#> - - - Processing information - - -
#> Loaded on Fri Sep 23 15:43:47 2016.
#> Normalised to sum of intensities.
#> Added markers from 'mrk' marker vector. Fri Sep 23 15:43:47 2016
#> MSnbase version: 1.99.2
# }