Becoming a better scientist with open and reproducible research
This blog post summarises the notes for my talk at the Are you ready for publishing reproducible research? meeting at the TU Delft on the 16 May 2019. The slides are available here.
This material is available under CC-BY, unless otherwise stated.
The original title of the talk was Being a better scientist with open and reproducible research, but I feel that this is some one constantly aims for, hence the change to becoming one.
Disclaimer: I do not speak from authority1. I speak of personal experience. My experience is in computational biology, bioinformatics and high throughput biology data. My experience doesn’t directly translate to other fields or domains (for example when it comes to data privacy) or even to other personalities in the same field.
Note 1: A piece of open research doesn’t automatically make it good, where good is defined as of high academic quality. A piece of closed research doesn’t make it bad, where bad here is defined of low academic quality. So openness doesn’t equate to academic quality. But openness provides some desired quality (i.e. desirable property) independent from academic excellent. Openness leads to trust.
Note 2: A piece of reproducible research doesn’t automatically make it good, where good is defined as of high academic quality. A piece of non reproducible research doesn’t make it bad, where bad here is defined of low academic quality. So reproducible doesn’t equate to academic quality. But reproducibility provides some desired quality (i.e. desirable property) independent from academic excellent. Reproducibility leads (among other things) to trust.
Open and reproducible research
Open != reproducible
Open research and reproducible research aren’t the same thing, and one doesn’t imply the other. They are historically also very different.
Many people seem to think that #OpenScience & the reproducibility crisis in psychology are somehow causally related. They are not. Open science is decades old & did not focus on reproducibility as a single issue — more here: https://t.co/KpJHIEqPj3 & here: https://t.co/KdMeK6PCUT pic.twitter.com/qF5yPTqNqu
— Olivia Guest | Ολίβια Γκεστ (@o_guest) December 1, 2018
From a technical point of view:
- When individual patronage funded scientists, discoveries were kept private or publicised as codes in anagrams or cyphers (Source Wikipedia:Open Science).
- The concept of open access to scientific data was institutionally established with the formation of the World Data Center system (now the World Data System) in 1957. MEDLINE, later renamed PubMed, was created in 1966. (Source Wikipedia:Open Science Data).
From a philosophical point of view:
The Mertonian norms (1942)
-
Communism: all scientists should have common ownership of scientific goods (intellectual property), to promote collective collaboration; secrecy is the opposite of this norm.
-
Universalism: scientific validity is independent of the sociopolitical status/personal attributes of its participants.
-
Disinterestedness: scientific institutions act for the benefit of a common scientific enterprise, rather than for the personal gain of individuals within them
-
Organised scepticism: scientific claims should be exposed to critical scrutiny before being accepted: both in methodology and institutional codes of conduct.
There isn’t only one type of open science
Open science has seen a continuous evolution since the 17th century, with the advent of dissemination of research in scientific journals and the societal demand to access scientific knowledge at large. Technology and communication has further accelerated this evolution, and put it in the spot light among researchers and academics (for for examples funder mandates) and more widely in the press with the cost of publications (see for example this Guardian long read article Is the staggeringly profitable business of scientific publishing bad for science? or the Paywall movie).
Open science/research is the process of transparent dissemination and access to knowledge, that can be applied to various scientific practices (image below from Wikipedia):

As a result
Open science/research can mean different things to different people, in particular when declined it along its many technical and philosophical attributes.
Take home message:
Open isn’t binary, it’s a gradient, it’s multidisciplinary, it’s multidimensional.
How to be an open scientist:
Let’s be open and understanding of different situations and constraints.
Why becoming an open research practitioners
It’s the right thing to do. See the The Mertonian norms… Or is it?
Benefits for your academic career: some examples from the Open as a career boost paragraph:
- Open access articles get more citations.
- Data availability is associated with citation benefit.
- Openly available software more likely to be used. (I don’t have any reference for this, and there are of course many couter examples).
Networking opportunities (I’m here thanks to my open research activities with my former colleague Marta Teperek at the University of Cambridge, UK).
See also Why Open Research
-
Increase your visibility: Build a name for yourself. Share your work and make it more visible.
-
Reduce publishing costs: Open publishing can cost the same or less than traditional publishing.
-
Take back control: Know your rights. Keep your rights. Decide how your work is used
-
Publish where you want: Publish in the journal of your choice and archive an open copy. (See The cost of knowledge boycott of Elsevier).
-
Get more funding: Meet funder requirements, and qualify for special funds such as the Wellcome Trust Open Research Fund.
-
Get that promotion: Open research is increasingly recognised in promotion and tenure. See also Reproducibility and open science are starting to matter in tenure and promotion July 14th, 2017, Brian Nosek) and the EU’s Evaluation of Research Careers fully acknowledging Open Science Practice defines an Open Science Career Assessment Matrix (OS-CAM):
But are there any risks?
Does it take more time to work openly?
Isn’t it worth investing time is managing data in a way that others (including future self) can find and understand it? That’s, IMHO, particularly important from a group leader’s perspective, where I want to build a corpus of data/software/research that other lab members can find, mine and re-use.
Are senior academics always supportive?
No.
Is there a risk of being scooped?
There certainly is a benefit if releasing one’s research early!
But, importantly, working with open and reproducible research in mind doesn’t mean releasing everything prematurely, it means
-
managing research in a way one can find data and results at every stage
-
one can reproduce/repeat results, re-run/compare them with new data or different methods/parameters, and
-
one can release data (or parts thereof) when/if appropriate.
So, are there any risks?
The Bullied Into Bad Science campaign is an initiative by early career researchers (ECRs) for early career researchers who aim for a fairer, more open and ethical research and publication environment.

Why reproducibility is important
-
For scientific reasons: think reproducibility crisis.
-
For political reasons: public trust in science, in data, in experts; without (public) trust in science and research, there won’t be any funding anymore.
But what do we mean by reproducibility?
From a But what to we mean by reproducibility? blog post.
-
Repeat my experiment, i.e. obtain the same tables/graphs/results using the same setup (data, software, …) in the same lab or on the same computer. That’s basically re-running one of my analysis some time after I original developed it.
-
Reproduce an experiment (not mine), i.e. obtain the same tables/graphs/results in a different lab or on a different computer, using the same setup (the data would be downloaded from a public repository and the same software, but possibly different version, different OS, is used). I suppose, we should differentiate replication using a fresh install and a virtual machine or docker image that replicates the original setup.
-
Replicate an experiment, i.e. obtain the same (similar enough) tables/graphs/results in a different set up. The data could still be downloaded from the public repository, or possibly re-generate/re-simulate it, and the analysis would be re-implemented based on the original description. This requires openness, and one would clearly not be allowed the use a black box approach (VM, docker image) or just re-running a script.
-
Finally, re-use the information/knowledge from one experiment to run a different experiment with the aim to confirm results from scratch.
Another view (from a talk by Kirstie Whitaker):
| Same Data | Different Data | |
|---|---|---|
| Same Code | reproduce | replicate |
| Different Code | robust | generalisable |
See also this opinion piece by Jeffrey T. Leek and Roger D. Peng, Reproducible research can still be wrong: Adopting a prevention approach.
From
Gabriel Becker An Imperfect Guide to Imperfect Reproducibility, May Institute for Computational Proteomics, 2019.
(Computational) Reproducibility Is Not The Point
Take home message:
The goal is trust, verification and guarantees:
- Trust in Reporting - result is accurately reported
- Trust in Implementation - analysis code successfully implements chosen methods
- Statistical Trust - data and methods are (still) appropriate
- Scientific Trust - result convincingly supports claim(s) about underlying systems or truths
Reproducibility As A Trust Scale (copyright Genentech Inc)
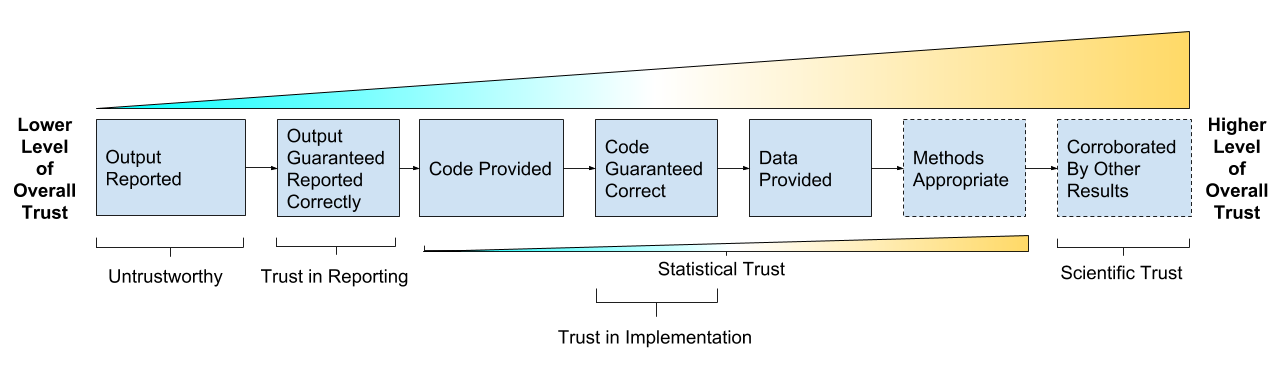
Take home message:
Reproducibility isn’t binary, it’s a gradient, it’s multidisciplinary, it’s multidimensional.
Another take home message:
Reproducibility isn’t easy.
Why becoming a reproducible research practitioners
Florian Markowetz, Five selfish reasons to work reproducibly, Genome Biology 2015, 16:274.
And so, my fellow scientists: ask not what you can do for reproducibility; ask what reproducibility can do for you! Here, I present five reasons why working reproducibly pays off in the long run and is in the self-interest of every ambitious, career-oriented scientist.
- Reason number 1: reproducibility helps to avoid disaster
- Reason number 2: reproducibility makes it easier to write papers
- Reason number 3: reproducibility helps reviewers see it your way
- Reason number 4: reproducibility enables continuity of your work
- Reason number 5: reproducibility helps to build your reputation
And career perspectives: Faculty promotion must assess reproducibility.
What can you do to improve trust in (your) research?
- Be an open research practitioners
- Be an reproducible research practitioners
Includes (but not limited to)
Preprints are the best!
Read, post, review and cite preprints (see ASAPbio for lots of resources about preprints).
Promoting open research through peer review
This section is based on my The role of peer-reviewers in checking
supporting information promoting open
science
talk.
As an open researcher, I think it is important to apply and promote the importance of data and good data management on a day-to-day basis (see for example Marta Teperek’s 2017 Data Management: Why would I bother? slides), but also to express this ethic in our academic capacity, such as peer review. My responsibility as a reviewer is to
- Accept sound/valid research and provide constructive comments
and hence
- Focus firstly on the validity of the research by inspecting the data, software and method. If the methods and/or data fail, the rest is meaningless.
I don’t see novelty, relevance, news-worthiness as my business as a reviewer. These factors are not the prime qualities of thorough research, but rather characteristics of flashy news.
Here are some aspects that are easy enough to check, and go a long way to verify the availability and validity and of the data
-
Availability: Are the data/software/methods accessible and understandable in a way that would allow an informed researcher in the same or close field to reproduce and/or verify the results underlying the claims? Note that this doesn’t mean that as a reviewer, I will necessarily try to repeat the whole analysis (that would be too time consuming indeed). But, conversely, a submission without data/software will be reviewed (and rejected, or more appropriately send back for completion) in matters of minutes. Are the data available in a public repository that guarantees that it will remain accessible, such as a subject-specific or, if none is available, a generic repository (such as zenodo or figshare, …), an institutional repository, or, but less desirable, supplementary information or a personal webpage2.
-
Meta-data: It’s of course not enough to provide a wild dump of the data/software/…, but these need to be appropriately documented. Personally, I recommend an
READMEfile in every top project directory to summarise the project, the data, … -
Do numbers match?: The first thing when reproducing someone’s analysis is to match the data files to the experimental design. That is one of the first things I check when reviewing a paper. For example if the experimental design says there are 2 groups, each with 3 replicates, I expect to find 6 (or a multiple thereof) data files or data columns in the data matrix. Along these lines, I also look at the file names (of column names in the data matrix) for a consistent naming convention, that allows to match the files (columns) to the experimental groups and replicates.
-
What data, what format: Is the data readable with standard and open/free software? Are the raw and processed available, and have the authors described how to get from one to the other?
-
License: Is the data shared in a way that allows users to re-use it. Under what conditions? Is the research output shared under a valid license?
Make sure that the data adhere to the FAIR principles:
Findable and Accessible and Interoperable and Reusable
Note that SI are not FAIR, not discoverable, not structured, voluntary, used to bury stuff. A personal web page is likely to disappear in the near future.
As a quick note, my ideal review system would be one where
-
Submit your data to a repository, where it gets checked (by specialists, data scientists, data curators) for quality, annotation, meta-data.
-
Submit your research with a link to the peer reviewed data. First review the intro and methods, then only the results (to avoid positive results bias).
When talking about open research and peer review, one logical extension is open peer review.
While I personally value open peer review and practice it when possible, it can be a difficult issue for ECRs, exposing them unnecessarily when reviewing work from prominent peers. It also can reinforce an already unwelcoming environment for underrepresented minorities. See more about this in the Inclusivity: open science and open science section below.
Registered reports
Define you data collection and analysis protocol in advance. Get it reviewed and, if accepted, get right to publish once data have been collected and analysed, irrespective of the (positive or negative) result.
-
Three challenges: Restrictions on flexibility (no p-hacking of HARKing), The time cost, Incentive structure isn’t in place yet.
-
Three benefits: Greater faith in research (no p-hacking of HARKing), New helpful systems (see technical solutions below), Investment in your future.
See https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000246 (2019).
Make allies
This is very important!
- Other ECR
- Librarians
- Data stewards/champions
- Research Software engineers
- On/off-line networking
Open research can lead to collaborative research. The
development of MSnbase is an
example I am very proud
of.
The credit goes to the outstanding contributions and contributors!
— Laurent Gⓐtt⓪ (@lgatt0) May 6, 2019
Collaborative work and cooperation is certainly one important concept that gravitates around open science/research (see the Mertonian norm of communism), but that isn’t necessary nor sufficient for open science.
Just do it!
Build openness at the core your research
(according to you possibilities)
Open and reproducible research doesn’t work if it’s an afterthought.
Technical solutions
-
Scripting, scripting, scripting (applies to code, data, analyses, manuscripts, …).
-
Avoid manual steps.
-
Document everything, especially manual steps (which you should avoid anyway).
-
Version control, such as git/github, bitbucket, …
-
Literate analyses: reproducible documents with R markdown, Sweave (R with LaTeX), Juyter notebooks, …
-
Shareable compute environments (docker containers).
-
Document and share all artefacts related to your research (when possible): data, code, protocols, …
See also An Imperfect Guide to Imperfect Reproducibility for further details.
Some examples of my own research
Spatial proteomics software: systematic and high throughput analysis of sub-cellular protein localisation.
Software
-
Software: infrastructure with
MSnbase(Gatto and Lilley 2012); dedicated machine learning withpRoloc(Gatto et al. 2014); interactive withpRolocGUI; data withpRolocdata(Gatto et al. 2014). -
The Bioconductor project (Huber et al. 2015) ecosystem for high throughput biology data analysis and comprehension: open source, and coordinated and collaborative (between and within domains/software) open development, enabling reproducible research, enables understanding of the data (not a black box) and drive scientific innovation.
QSep: quantify resolution of a spatial proteomics experiment
QSep is a function within the pRoloc software.
Evolution of Gatto et al. (2018)
- left: reproducible document
- centre: preprint
- right: peer reviewer paper
SpatialMap
SpatialMap is a project aiming at producing a visualisation and data
sharing platform for spatial proteomics. I decided to promote and
drive it as openly as possible in the frame of the Open Research
Pilot
Project. The
ORPP is a joint project by the Office of Scholarly
Communication at the University of Cambridge
and the Wellcome Trust Open research
team. Here
are the reasons why the SpatialMap project is an open project:
-
The SpatialMap project in itself is about opening up spatial proteomics data by facilitating data sharing and providing tools to further the comprehension of the data. One aim is to allow users to use the SpatialMap web portal to upload, share, explore and discuss their data privately with collaborators in a first instance (few researchers share their data before publication), then make the data available to reviewer, and finally, once reviewed, make it public at the push of a button. The incentive for early utilisation of the platform is to provide interactive data visualisation and integration with other tools and sharing of the data with close collaborators.
-
The project is developed completely openly in a public GitHub repository. Absolutely all code and contributions are publicly available. Anyone can collaborate, or even fork the project and build their own.
-
I publicly announced the SpatialMap project in a blog post. The blog post was written as a legitimate grant application (albeit a little bit shorter and sticking to the most important parts).
Note that I do not have any dedicated funding for this project. The progress so far was the result of a masters student visiting my group, and is currently not actively developed anymore.
Inclusivity: open research and open research
There is
Open Science as in widely disseminated and openly accessible
and
Open Science as in inclusive and welcoming
On being inclusive - Twitter thread by Cameron Neylon:
The primary value proposition of #openscience is that diverse contributions allow better critique, refinement, and application 3/n
— CⓐmeronNeylon (@CameronNeylon) August 10, 2017
It was a damned hard community to break into. Any step I took to be more open, I felt attacked for not doing enough/doing it right.
— Christie Bahlai (@cbahlai) June 4, 2017
As far as I was concerned for a long time (until June 2017 to be accurate - this section is based this Open science and open science post), the former more technical definition was always what I was focusing on, and the second community-level aspect of openness was, somehow, implicit from the former, but that’s clearly not the case.
Even if there are efforts to promote diversity, under-represented minorities (URM) don’t necessarily feel included. When it comes to open science/research URMs can be further discriminated against by greater exposure or, can’t always afford to be vocal.
- Not everybody has the privilege to be open.
- There are different levels in how open one wants, or how open one could afford to be.
- Every voice and support is welcome.
Conclusions
Standing on the shoulders of giants only really makes sense in the context of open and reproducible research.
-
If you are here (or have read this), chances are you are on the path towards open and reproducible research.
-
You are the architect of the kind of research and researcher you want to become. I hope these include openness and reproducibility.
-
It’s a long path, that constantly evolves, depending on constraints, aspirations, environment, …
-
The sky is the limit, be creative: work out the (open and reproducible) research that works for you now …
-
… and that you want to work (for you and others) in the future.
Acknowledgements
One of my advice was to make allies. I have been lucky to meet wonderful allies and inspiring friends along the path towards open and reproducible research that works for me. Among these, I would like to highlight Corina Logan, Stephen Eglen, Marta Teperek, Kirstie Whitaker, Chris Hartgenink, Naomie Penfold, Yvonne Nobis.
-
I actually think that authority (or seniority) isn’t doing any favours when it comes to open research and reproducibility. The more senior stakeholder all too often aren’t those that drive research toward more openness and reproducibility. There are, fortunately, notable exceptions. ↩
-
There is often no perfect solution, and a combination of the above might be desirable. ↩

