Chapter 13 R programming concepts and tools
This section is composed of various section of more advanced programming topics from the Teaching Material page.
13.1 Defensive programming
Before even debugging, let’s look at ways to prevent bugs in the first place.
Defensive programming:
- making the code work in a predicable manner
- writing code that fails in a well-defined manner
- if something weird happens, either properly deal with it, of fail quickly and loudly
The level of defensiveness will depend whether you write a function for interactive of programmatic usage.
Talking to users
Diagnostic messages
message("This is a message for our dear users.")message("This is a message for our dear users. ",
paste("Thank you for using our software",
sw, "version", packageVersion(sw)))Do not use print or cat:
f1 <- function() {
cat("I AM LOUD AND YOU CAN'T HELP IT.\n")
## do stuff
invisible(TRUE)
}
f1()f2 <- function() {
message("Sorry to interup, but...")
## do stuff
invisible(TRUE)
}
f2()
suppressMessages(f2())Of course, it is also possible to manually define verbosity. This makes you write more code for a feature readily available. But still better to use message.
f3 <- function(verbose = TRUE) {
if (verbose)
message("I am being verbose because you let me.")
## do stuff
invisible(TRUE)
}
f3()
f3(verbose = FALSE)Warning
There is a problem with warnings. No one reads them. Pat Burns, in R inferno.
warning("Do not ignore me. Somthing bad might have happened.")
warning("Do not ignore me. Somthing bad might be happening.", immediate. = TRUE)f <- function(...)
warning("Attention, attention, ...!", ...)
f()
f(call. = FALSE)Print warnings after they have been thrown.
warnings()
last.warningSee also to warn option in ?options .
option("warn")Error
stop("This is the end, my friend.")log(c(2, 1, 0, -1, 2)); print('end') ## warning
xor(c(TRUE, FALSE)); print ('end') ## errorStop also has a call. parameter.
geterrmessage()Progress bars
utils::txtProgressBarfunction
n <- 10
pb <- txtProgressBar(min = 0, max = n, style = 3)
for (i in 1:n) {
setTxtProgressBar(pb, i)
Sys.sleep(0.5)
}
close(pb)progresspackage
library("progress")
pb <- progress_bar$new(total = n)
for (i in 1:n) {
pb$tick()
Sys.sleep(0.5)
}Tip: do not over use progress bars. Ideally, a user should be confident that everything is under control and progress is made while waiting for a function to return. In my experience, a progress bar is usefull when there is a specific and/or user-defined number of iterations, such a iterating over n files, or running a simulation n times.
13.2 KISS
Keep your functions simple and stupid (and short).
13.3 Failing fast and well
Bounds errors are ugly, nasty things that should be stamped out whenever possible. One solution to this problem is to use the
assertstatement. Theassertstatement tells C++, “This can never happen, but if it does, abort the program in a nice way.” One thing you find out as you gain programming experience is that things that can “never happen” happen with alarming frequency. So just to make sure that things work as they are supposed to, it’s a good idea to put lots of self checks in your program. – Practical C++ Programming, Steve Oualline, O’Reilly.
if (!condition) stop(...)stopifnot(TRUE)
stopifnot(TRUE, FALSE)For example to test input classes, lengths, …
f <- function(x) {
stopifnot(is.numeric(x), length(x) == 1)
invisible(TRUE)
}
f(1)
f("1")
f(1:2)
f(letters)The assertthat package:
x <- "1"
library("assertthat")
stopifnot(is.numeric(x))
assert_that(is.numeric(x))
assert_that(length(x) == 2)assert_that()signal an error.see_if()returns a logical value, with the error message as an attribute.validate_that()returnsTRUEon success, otherwise returns the error as a string.is.flag(x): is xTRUEorFALSE? (a boolean flag)is.string(x): is x a length 1 character vector?has_name(x, nm),x %has_name% nm: doesxhave componentnm?has_attr(x, attr),x %has_attr% attr: doesxhave attributeattr?is.count(x): is x a single positive integer?are_equal(x, y): arexandyequal?not_empty(x): are all dimensions ofxgreater than 0?noNA(x): isxfree from missing values?is.dir(path): ispatha directory?is.writeable(path)/is.readable(path): ispathwriteable/readable?has_extension(path, extension): doesfilehave givenextension?
13.4 Consistency and predictability
Ineractive use vs programming: Moving from using R to programming R is abstraction, automation, generalisation.
drop
head(cars)
head(cars[, 1])
head(cars[, 1, drop = FALSE])sapply/lapply
df1 <- data.frame(x = 1:3, y = LETTERS[1:3])
sapply(df1, class)
df2 <- data.frame(x = 1:3, y = Sys.time() + 1:3)
sapply(df2, class)Rather use a form where the return data structure is known…
lapply(df1, class)
lapply(df2, class)or that will break if the result is not what is exected
vapply(df1, class, "1")
vapply(df2, class, "1")Reminder of the interactive use vs programming examples: - [ and drop - sapply, lapply, vapply
Remember also the concept of tidy data.
13.5 Comparisons
Floating point issues to be aware of
R FAQ 7.31?
a <- sqrt(2)
a * a == 2
a * a - 21L + 2L == 3L
1.0 + 2.0 == 3.0
0.1 + 0.2 == 0.3Floating point: how to compare
all.equalcompares R objects for near equality. Takes into account whether object attributes and names ought the taken into consideration (check.attributesandcheck.namesparameters) and tolerance, which is machine dependent.
all.equal(0.1 + 0.2, 0.3)
all.equal(0.1 + 0.2, 3.0)
isTRUE(all.equal(0.1 + 0.2, 3)) ## when you just want TRUE/FALSEExact identity
identical: test objects for exact equality
1 == NULL
all.equal(1, NULL)
identical(1, NULL)
identical(1, 1.) ## TRUE in R (both are stored as doubles)
all.equal(1, 1L)
identical(1, 1L) ## stored as different typesAppropriate within if, while condition statements. (not all.equal, unless wrapped in isTRUE).
13.6 Exercise
From Advanced R by Hadley Wickham.
The col_means function computes the means of all numeric columns in a data frame.
col_means <- function(df) {
numeric <- sapply(df, is.numeric)
numeric_cols <- df[, numeric]
data.frame(lapply(numeric_cols, mean))
}Is it a robust function? What happens if there are unusual inputs.
col_means(mtcars)
col_means(mtcars[, 0])
col_means(mtcars[0, ])
col_means(mtcars[, "mpg", drop = FALSE])
col_means(1:10)
col_means(as.matrix(mtcars))
col_means(as.list(mtcars))
mtcars2 <- mtcars
mtcars2[-1] <- lapply(mtcars2[-1], as.character)
col_means(mtcars2)Using some of the concepts and tips above, re-write col_means to make it more robust.
13.7 Debugging: techniques and tools
Shit happens!
Funding your bug is a process of confirming the many things that you believe are true - until you find one which is not true. – Norm Matloff
1. Identify the bug (the difficult part)
- Something went wrong!
- Where in the code does it happen?
- Does it happen every time?
- What input triggered it?
- Report it (even if it is in your code - use github issues, for example).
Tip: Beware of your intuition. As a scientist, do what you are used to: generate a hypotheses, design an experiment to test them, and record the results.
2. Fix it (the less difficult part)
- Correct the bug.
- Make sure that bug will not repeat itself!
- How can we be confident that we haven’t introduced new bugs?
Tools
print/cattraceback()browser()- IDE: RStudio, StatET, emacs’ ess tracebug.
Manually
Inserting print and cat statements in the code. Works, but time consuming.
Finding the bug
Bugs are shy, and are generally hidden, deep down in your code, to make it as difficult as possible for you to find them.
e <- function(i) {
x <- 1:4
if (i < 5) x[1:2]
else x[-1:2]
}
f <- function() sapply(1:10, e)
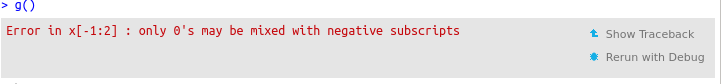
g <- function() f()traceback: lists the sequence of calls that lead to the error
g()
traceback()If the source code is available (for example for source()d code), then traceback will display the exact location in the function, in the form filename.R#linenum.
Browsing the error
Register the function for debugging:
debug(g). This adds a call to thebrowser()function (see also below) and the very beginning of the functiong.Every call to
g()will not be run interactively.To finish debugging:
undebug(g).
debug(g)
g()How to debug:
nexecutes the next step of the function. Useprint(n)orget(n)to print/access the variablen.sto step into the next function. If it is not a function, same asn.fto finish execution of the current loop of function.cto leave interactive debugging and continue regular execution of the function.Qto stop debugging, terminate the function and return to the global workspace.whereprint a stack trace of all active function calls.Entersame asn(ors, if it was used most recently), unlessoptions(browserNLdisabled = TRUE)is set.
To fix a function when the source code is not directly available, use fix(fun). This will open the function’s source code for editing and, after saving and closing, store the updated function in the global workspace.
Breakpoints
Add a call to
browser()anywhere in the source code to execute the rest of the code interactively.To run breakpoints conditionally, wrap the call to
browser()in a condition.
Debugging with IDEs
- RSudio:
Show Traceback,Rerun with Debugand interactive debugging.
StatET (Eclipse plugin)
Exercise
- Your turn - play with
traceback,recoveranddebug:
(Example originally by Martin Morgan and Robert Gentleman.)
e <- function(i) {
x <- 1:4
if (i < 5) x[1:2]
else x[-1:2] # oops! x[-(1:2)]
}
f <- function() sapply(1:10, e)
g <- function() f()- Fix
readFasta2.
Preparing the ground
## make sure you have the 'sequences' package.
library("devtools")
install_github("lgatto/sequences") ## from github
## or
install.packages("sequences") ## from CRANA working example: reading a single sequence from a fasta file to create a object of class DnaSeq, representing the DNA string:
library("sequences")## Loading required package: Rcpp## This is package 'sequences'##
## Attaching package: 'sequences'## The following object is masked from 'package:dplyr':
##
## idf <- dir(system.file("extdata", package = "sequences"),
full.names=TRUE, pattern = "aDnaSeq.fasta")
readFasta(f)## Object of class DnaSeq
## Id: example dna sequence
## Length: 132
## Alphabet: A C G T
## Sequence: AGCATACGACGACTACGACACTACGACATCAGACACTACAGACTACTACGACTACAGACATCAGACACTACATATTTACATCATCAGAGATTATATTAACATCAGACATCGACACATCATCATCAGCATCATA bug, trying to read multiple sequences from a fasta file. The expected behaviour would be to return a list of DnaSeq objects:
## Get readFasta2, the function to debug
sequences:::debugme()
## Get an example file
f <- dir(system.file("extdata", package = "sequences"),
full.names=TRUE, pattern = "moreDnaSeqs.fasta")
## BANG!
readFasta2(f)